Related Research Articles

Chromosomal crossover, or crossing over, is the exchange of genetic material during sexual reproduction between two homologous chromosomes' non-sister chromatids that results in recombinant chromosomes. It is one of the final phases of genetic recombination, which occurs in the pachytene stage of prophase I of meiosis during a process called synapsis. Synapsis begins before the synaptonemal complex develops and is not completed until near the end of prophase I. Crossover usually occurs when matching regions on matching chromosomes break and then reconnect to the other chromosome.

Exodeoxyribonuclease V is an enzyme of E. coli that initiates recombinational repair from potentially lethal double strand breaks in DNA which may result from ionizing radiation, replication errors, endonucleases, oxidative damage, and a host of other factors. The RecBCD enzyme is both a helicase that unwinds, or separates the strands of DNA, and a nuclease that makes single-stranded nicks in DNA. It catalyses exonucleolytic cleavage in either 5′- to 3′- or 3′- to 5′-direction to yield 5′-phosphooligonucleotides.

RuvABC is a complex of three proteins that mediate branch migration and resolve the Holliday junction created during homologous recombination in bacteria. As such, RuvABC is critical to bacterial DNA repair.

Homologous recombination is a type of genetic recombination in which genetic information is exchanged between two similar or identical molecules of double-stranded or single-stranded nucleic acids.
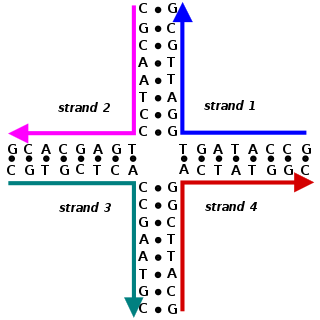
A Holliday junction is a branched nucleic acid structure that contains four double-stranded arms joined. These arms may adopt one of several conformations depending on buffer salt concentrations and the sequence of nucleobases closest to the junction. The structure is named after Robin Holliday, the molecular biologist who proposed its existence in 1964.

DNA mismatch repair protein Mlh1 or MutL protein homolog 1 is a protein that in humans is encoded by the MLH1 gene located on chromosome 3. It is a gene commonly associated with hereditary nonpolyposis colorectal cancer. Orthologs of human MLH1 have also been studied in other organisms including mouse and the budding yeast Saccharomyces cerevisiae.
Robin Holliday was a British molecular biologist. Holliday described a mechanism of DNA-strand exchange that attempted to explain gene-conversion events that occur during meiosis in fungi. That model first proposed in 1964 and is now known as the Holliday Junction.
Chromosome segregation is the process in eukaryotes by which two sister chromatids formed as a consequence of DNA replication, or paired homologous chromosomes, separate from each other and migrate to opposite poles of the nucleus. This segregation process occurs during both mitosis and meiosis. Chromosome segregation also occurs in prokaryotes. However, in contrast to eukaryotic chromosome segregation, replication and segregation are not temporally separated. Instead segregation occurs progressively following replication.

Bloom syndrome protein is a protein that in humans is encoded by the BLM gene and is not expressed in Bloom syndrome.

DNA mismatch repair protein Mlh3 is a protein that in humans is encoded by the MLH3 gene.

Junctional diversity describes the DNA sequence variations introduced by the improper joining of gene segments during the process of V(D)J recombination. This process of V(D)J recombination has vital roles for the vertebrate immune system, as it is able to generate a huge repertoire of different T-cell receptor (TCR) and immunoglobulin molecules required for pathogen antigen recognition by T-cells and B cells, respectively.

RecQ-mediated genome instability protein 1 is a protein that in humans is encoded by the RMI1 gene.
A Chi site or Chi sequence is a short stretch of DNA in the genome of a bacterium near which homologous recombination is more likely to occur than on average across the genome. Chi sites serve as stimulators of DNA double-strand break repair in bacteria, which can arise from radiation or chemical treatments, or result from replication fork breakage during DNA replication. The sequence of the Chi site is unique to each group of closely related organisms; in E. coli and other enteric bacteria, such as Salmonella, the core sequence is 5'-GCTGGTGG-3' plus important nucleotides about 4 to 7 nucleotides to the 3' side of the core sequence. The existence of Chi sites was originally discovered in the genome of bacteriophage lambda, a virus that infects E. coli, but is now known to occur about 1000 times in the E. coli genome.
The RecF pathway, also called the RecFOR pathway, is a pathway of homologous recombination that repairs DNA in bacteria. It repairs breaks that occur on only one of DNA's two strands, known as single-strand gaps. The RecF pathway can also repair double-strand breaks in DNA when the RecBCD pathway, another pathway of homologous recombination in bacteria, is inactivated by mutations. Like the RecBCD pathway, the RecF pathway requires RecA for strand invasion. The two pathways are also similar in their phases of branch migration, in which the Holliday junction slides in one direction, and resolution, in which the Holliday junctions are cleaved apart by enzymes.

Stephen Craig West FRS is a British biochemist and molecular biologist specialising in research on DNA recombination and repair. He is known for pioneering studies on genome instability diseases including cancer. West obtained his BSc in 1974, and his PhD in 1977, both from Newcastle University. He is currently a Principal Group Leader at the Francis Crick Institute in London. He is an honorary Professor at University College London, and at Imperial College London. In recognition of his work he was awarded the Louis-Jeantet Prize for Medicine in 2007, is a fellow of the Royal Society, the Academy of Medical Sciences, an International Member of the National Academy of Sciences, and an International Honorary Member of the American Academy of Arts and Sciences. He received the 2022 Royal Medal for 'discovering and determining the functions of key enzymes that are essential for DNA recombination, repair and the maintenance of genomes'.
MutS is a mismatch DNA repair protein, originally described in Escherichia coli.
Crossover junction endodeoxyribonuclease, also known as Holliday junction resolvase, Holliday junction endonuclease, Holliday junction-cleaving endonuclease, Holliday junction-resolving endoribonuclease, crossover junction endoribonuclease, and cruciform-cutting endonuclease, is an enzyme involved in DNA repair and homologous recombination. Specifically, it performs endonucleolytic cleavage that results in single-stranded crossover between two homologous DNA molecules at the Holliday junction to produce recombinant DNA products for chromosomal segregation. This process is known as Holliday junction resolution.

SLX4 interacting protein is a protein that in humans is encoded by the SLX4IP gene.
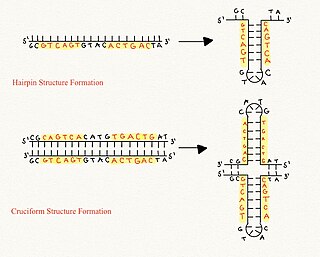
Cruciform DNA is a form of non-B DNA, or an alternative DNA structure. The formation of cruciform DNA requires the presence of palindromes called inverted repeat sequences. These inverted repeats contain a sequence of DNA in one strand that is repeated in the opposite direction on the other strand. As a result, inverted repeats are self-complementary and can give rise to structures such as hairpins and cruciforms. Cruciform DNA structures require at least a six nucleotide sequence of inverted repeats to form a structure consisting of a stem, branch point and loop in the shape of a cruciform, stabilized by negative DNA supercoiling.

GEN1, Holliday junction 5' flap endonuclease is a protein that in humans is encoded by the GEN1 gene.
References
- ↑ "Entrez Gene: Holliday junction recognition protein" . Retrieved 2013-05-17.