
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, responding to stimuli, providing structure to cells and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific 3D structure that determines its activity.

Proteomics is the large-scale study of proteins. Proteins are vital macromolecules of all living organisms, with many functions such as the formation of structural fibers of muscle tissue, enzymatic digestion of food, or synthesis and replication of DNA. In addition, other kinds of proteins include antibodies that protect an organism from infection, and hormones that send important signals throughout the body.

Centrifugation is a mechanical process which involves the use of the centrifugal force to separate particles from a solution according to their size, shape, density, medium viscosity and rotor speed. The denser components of the mixture migrate away from the axis of the centrifuge, while the less dense components of the mixture migrate towards the axis. Chemists and biologists may increase the effective gravitational force of the test tube so that the precipitate (pellet) will travel quickly and fully to the bottom of the tube. The remaining liquid that lies above the precipitate is called a supernatant or supernate.
Lysis is the breaking down of the membrane of a cell, often by viral, enzymic, or osmotic mechanisms that compromise its integrity. A fluid containing the contents of lysed cells is called a lysate. In molecular biology, biochemistry, and cell biology laboratories, cell cultures may be subjected to lysis in the process of purifying their components, as in protein purification, DNA extraction, RNA extraction, or in purifying organelles.
A lysis buffer is a buffer solution used for the purpose of breaking open cells for use in molecular biology experiments that analyze the labile macromolecules of the cells. Most lysis buffers contain buffering salts and ionic salts to regulate the pH and osmolarity of the lysate. Sometimes detergents are added to break up membrane structures. For lysis buffers targeted at protein extraction, protease inhibitors are often included, and in difficult cases may be almost required. Lysis buffers can be used on both animal and plant tissue cells.
In cell biology, cell fractionation is the process used to separate cellular components while preserving individual functions of each component. This is a method that was originally used to demonstrate the cellular location of various biochemical processes. Other uses of subcellular fractionation is to provide an enriched source of a protein for further purification, and facilitate the diagnosis of various disease states.

In biochemistry and cell biology, differential centrifugation is a common procedure used to separate organelles and other sub-cellular particles based on their sedimentation rate. Although often applied in biological analysis, differential centrifugation is a general technique also suitable for crude purification of non-living suspended particles. In a typical case where differential centrifugation is used to analyze cell-biological phenomena, a tissue sample is first lysed to break the cell membranes and release the organelles and cytosol. The lysate is then subjected to repeated centrifugations, where particles that sediment sufficiently quickly at a given centrifugal force for a given time form a compact "pellet" at the bottom of the centrifugation tube.
Protein purification is a series of processes intended to isolate one or a few proteins from a complex mixture, usually cells, tissues or whole organisms. Protein purification is vital for the specification of the function, structure and interactions of the protein of interest. The purification process may separate the protein and non-protein parts of the mixture, and finally separate the desired protein from all other proteins. Ideally, to study a protein of interest, it must be separated from other components of the cell so that contaminants will not interfere in the examination of the protein of interest's structure and function. Separation of one protein from all others is typically the most laborious aspect of protein purification. Separation steps usually exploit differences in protein size, physico-chemical properties, binding affinity and biological activity. The pure result may be termed protein isolate.
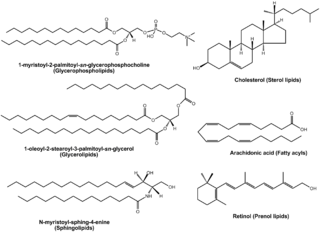
Lipidomics is the large-scale study of pathways and networks of cellular lipids in biological systems The word "lipidome" is used to describe the complete lipid profile within a cell, tissue, organism, or ecosystem and is a subset of the "metabolome" which also includes other major classes of biological molecules. Lipidomics is a relatively recent research field that has been driven by rapid advances in technologies such as mass spectrometry (MS), nuclear magnetic resonance (NMR) spectroscopy, fluorescence spectroscopy, dual polarisation interferometry and computational methods, coupled with the recognition of the role of lipids in many metabolic diseases such as obesity, atherosclerosis, stroke, hypertension and diabetes. This rapidly expanding field complements the huge progress made in genomics and proteomics, all of which constitute the family of systems biology.
The first isolation of deoxyribonucleic acid (DNA) was done in 1869 by Friedrich Miescher. DNA extraction is the process of isolating DNA from the cells of an organism isolated from a sample, typically a biological sample such as blood, saliva, or tissue. It involves breaking open the cells, removing proteins and other contaminants, and purifying the DNA so that it is free of other cellular components. The purified DNA can then be used for downstream applications such as PCR, sequencing, or cloning. Currently, it is a routine procedure in molecular biology or forensic analyses.
The transcriptome is the set of all RNA transcripts, including coding and non-coding, in an individual or a population of cells. The term can also sometimes be used to refer to all RNAs, or just mRNA, depending on the particular experiment. The term transcriptome is a portmanteau of the words transcript and genome; it is associated with the process of transcript production during the biological process of transcription.

Cell disruption is a method or process for releasing biological molecules from inside a cell.
A protein microarray is a high-throughput method used to track the interactions and activities of proteins, and to determine their function, and determining function on a large scale. Its main advantage lies in the fact that large numbers of proteins can be tracked in parallel. The chip consists of a support surface such as a glass slide, nitrocellulose membrane, bead, or microtitre plate, to which an array of capture proteins is bound. Probe molecules, typically labeled with a fluorescent dye, are added to the array. Any reaction between the probe and the immobilised protein emits a fluorescent signal that is read by a laser scanner. Protein microarrays are rapid, automated, economical, and highly sensitive, consuming small quantities of samples and reagents. The concept and methodology of protein microarrays was first introduced and illustrated in antibody microarrays in 1983 in a scientific publication and a series of patents. The high-throughput technology behind the protein microarray was relatively easy to develop since it is based on the technology developed for DNA microarrays, which have become the most widely used microarrays.

Fast protein liquid chromatography (FPLC) is a form of liquid chromatography that is often used to analyze or purify mixtures of proteins. As in other forms of chromatography, separation is possible because the different components of a mixture have different affinities for two materials, a moving fluid and a porous solid. In FPLC the mobile phase is an aqueous buffer solution. The buffer flow rate is controlled by a positive-displacement pump and is normally kept constant, while the composition of the buffer can be varied by drawing fluids in different proportions from two or more external reservoirs. The stationary phase is a resin composed of beads, usually of cross-linked agarose, packed into a cylindrical glass or plastic column. FPLC resins are available in a wide range of bead sizes and surface ligands depending on the application.

Quantitative proteomics is an analytical chemistry technique for determining the amount of proteins in a sample. The methods for protein identification are identical to those used in general proteomics, but include quantification as an additional dimension. Rather than just providing lists of proteins identified in a certain sample, quantitative proteomics yields information about the physiological differences between two biological samples. For example, this approach can be used to compare samples from healthy and diseased patients. Quantitative proteomics is mainly performed by two-dimensional gel electrophoresis (2-DE), preparative native PAGE, or mass spectrometry (MS). However, a recent developed method of quantitative dot blot (QDB) analysis is able to measure both the absolute and relative quantity of an individual proteins in the sample in high throughput format, thus open a new direction for proteomic research. In contrast to 2-DE, which requires MS for the downstream protein identification, MS technology can identify and quantify the changes.
RNA extraction is the purification of RNA from biological samples. This procedure is complicated by the ubiquitous presence of ribonuclease enzymes in cells and tissues, which can rapidly degrade RNA. Several methods are used in molecular biology to isolate RNA from samples, the most common of these is guanidinium thiocyanate-phenol-chloroform extraction. The filter paper based lysis and elution method features high throughput capacity.
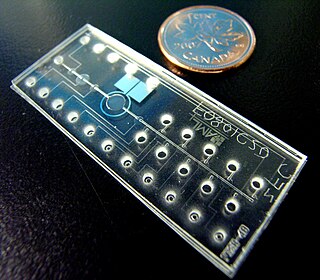
Bio-MEMS is an abbreviation for biomedical microelectromechanical systems. Bio-MEMS have considerable overlap, and is sometimes considered synonymous, with lab-on-a-chip (LOC) and micro total analysis systems (μTAS). Bio-MEMS is typically more focused on mechanical parts and microfabrication technologies made suitable for biological applications. On the other hand, lab-on-a-chip is concerned with miniaturization and integration of laboratory processes and experiments into single chips. In this definition, lab-on-a-chip devices do not strictly have biological applications, although most do or are amenable to be adapted for biological purposes. Similarly, micro total analysis systems may not have biological applications in mind, and are usually dedicated to chemical analysis. A broad definition for bio-MEMS can be used to refer to the science and technology of operating at the microscale for biological and biomedical applications, which may or may not include any electronic or mechanical functions. The interdisciplinary nature of bio-MEMS combines material sciences, clinical sciences, medicine, surgery, electrical engineering, mechanical engineering, optical engineering, chemical engineering, and biomedical engineering. Some of its major applications include genomics, proteomics, molecular diagnostics, point-of-care diagnostics, tissue engineering, single cell analysis and implantable microdevices.

Laser ablation electrospray ionization (LAESI) is an ambient ionization method for mass spectrometry that combines laser ablation from a mid-infrared (mid-IR) laser with a secondary electrospray ionization (ESI) process. The mid-IR laser is used to generate gas phase particles which are then ionized through interactions with charged droplets from the ESI source. LAESI was developed in Professor Akos Vertes lab by Peter Nemes in 2007 and it was marketed commercially by Protea Biosciences, Inc until 2017. Fiber-LAESI for single-cell analysis approach was developed by Bindesh Shrestha in Professor Vertes lab in 2009. LAESI is a novel ionization source for mass spectrometry (MS) that has been used to perform MS imaging of plants, tissues, cell pellets, and even single cells. In addition, LAESI has been used to analyze historic documents and untreated biofluids such as urine and blood. The technique of LAESI is performed at atmospheric pressure and therefore overcomes many of the obstacles of traditional MS techniques, including extensive and invasive sample preparation steps and the use of high vacuum. Because molecules and aerosols are ionized by interacting with an electrospray plume, LAESI's ionization mechanism is similar to SESI and EESI techniques.
Single cell oil, also known as Microbial oil consists of the intracellular storage lipids, triacyglycerols. It is similar to vegetable oil, another biologically produced oil. They are produced by oleaginous microorganisms, which is the term for those bacteria, molds, algae and yeast, which can accumulate 20% to 80% lipids of their biomass. The accumulation of lipids take place by the end of logarithmic phase and continues during station phase until carbon source begins to reduce with nutrition limitation.

In the field of cellular biology, single-cell analysis and subcellular analysis is the study of genomics, transcriptomics, proteomics, metabolomics and cell–cell interactions at the single cell level. The concept of single-cell analysis originated in the 1970s. Before the discovery of heterogeneity, single-cell analysis mainly referred to the analysis or manipulation of an individual cell in a bulk population of cells at a particular condition using optical or electronic microscope. To date, due to the heterogeneity seen in both eukaryotic and prokaryotic cell populations, analyzing a single cell makes it possible to discover mechanisms not seen when studying a bulk population of cells. Technologies such as fluorescence-activated cell sorting (FACS) allow the precise isolation of selected single cells from complex samples, while high throughput single cell partitioning technologies, enable the simultaneous molecular analysis of hundreds or thousands of single unsorted cells; this is particularly useful for the analysis of transcriptome variation in genotypically identical cells, allowing the definition of otherwise undetectable cell subtypes. The development of new technologies is increasing our ability to analyze the genome and transcriptome of single cells, as well as to quantify their proteome and metabolome. Mass spectrometry techniques have become important analytical tools for proteomic and metabolomic analysis of single cells. Recent advances have enabled quantifying thousands of protein across hundreds of single cells, and thus make possible new types of analysis. In situ sequencing and fluorescence in situ hybridization (FISH) do not require that cells be isolated and are increasingly being used for analysis of tissues.











