Heat shock proteins (HSP) are a family of proteins that are produced by cells in response to exposure to stressful conditions. They were first described in relation to heat shock, but are now known to also be expressed during other stresses including exposure to cold, UV light and during wound healing or tissue remodeling. Many members of this group perform chaperone functions by stabilizing new proteins to ensure correct folding or by helping to refold proteins that were damaged by the cell stress. This increase in expression is transcriptionally regulated. The dramatic upregulation of the heat shock proteins is a key part of the heat shock response and is induced primarily by heat shock factor (HSF). HSPs are found in virtually all living organisms, from bacteria to humans.
The 5′ untranslated region is the region of an mRNA that is directly upstream from the initiation codon. This region is important for the regulation of translation of a transcript by differing mechanisms in viruses, prokaryotes and eukaryotes. While called untranslated, the 5′ UTR or a portion of it is sometimes translated into a protein product. This product can then regulate the translation of the main coding sequence of the mRNA. In many organisms, however, the 5′ UTR is completely untranslated, instead forming complex secondary structure to regulate translation.
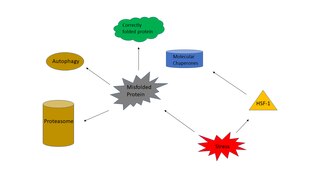
The heat shock response (HSR) is a cell stress response that increases the number of molecular chaperones to combat the negative effects on proteins caused by stressors such as increased temperatures, oxidative stress, and heavy metals. In a normal cell, proteostasis must be maintained because proteins are the main functional units of the cell. Many proteins take on a defined configuration in a process known as protein folding in order to perform their biological functions. If these structures are altered, critical processes could be affected, leading to cell damage or death. The heat shock response can be employed under stress to induce the expression of heat shock proteins (HSP), many of which are molecular chaperones, that help prevent or reverse protein misfolding and provide an environment for proper folding.

The apolipoprotein B (apoB) 5′ UTR cis regulatory element is an RNA element located in the 5′ UTR of the human apoB mRNA. This structured element increases translation of the apoB protein or a reporter gene.

The BiP internal ribosome entry site (IRES) is an RNA element present in the 5' UTR of the mRNA of BiP protein and allows cap-independent translation. BiP protein expression has been found to be significantly enhanced by the heat shock response due to internal ribosome entry site (IRES)-dependent translation. It is thought that this translational mechanism is essential for the survival of cells under stress.
The heat shock protein 70 (Hsp70) internal ribosome entry site (IRES) is an RNA element that allows cap independent translation during conditions such as heat shock and stress. It has been shown that the 216 nucleotide long 5' UTR contains internal ribosome entry site activity.

Nanos 3′ UTR translation control element is a cis-regulatory element in the 3′ untranslated region of the messenger RNA which encodes the Nanos protein. The Nanos protein in Drosophila is required for correct morphogenesis in the Drosophila embryo. Translation of the Nanos mRNA is repressed in the bulk cytoplasm and activated in the posterior region. The translation control element (TCE) in the 3'UTR forms a Y-shaped secondary structure, part of which is recognised by the Smaug protein and leads to translational repression.

The repression of heat shock gene expression (ROSE) element is an RNA element found in the 5' UTR of some heat shock protein's mRNAs. The ROSE element is an RNA thermometer that negatively regulates heat shock gene expression. The secondary structure is thought to be altered by temperature, thus it is an RNA thermometer. This structure blocks access to the ribosome binding site at normal temperatures. During heat shock however, the structure changes freeing the ribosome binding site and allowing expression to occur.

mRNA localization is a common mode of posttranscriptional regulation of gene expression that targets a protein to its site of function. Proteins are highly dependent on cellular environments for stability and function, therefore, mRNA localization signals are crucial for maintaining protein function. The Gurken localisation signal is an RNA regulatory element conserved across many species of Drosophila. The element consists of an RNA stem loop within the coding region of the messenger RNA that forms a signal for dynein-mediated Gurken mRNA transport to the dorsoanterior cap near the nucleus of the oocyte.

Heat shock protein HSP 90-alpha is a protein that in humans is encoded by the HSP90AA1 gene.
Trans-regulatory elements (TRE) are DNA sequences encoding upstream regulators, which may modify or regulate the expression of distant genes. Trans-acting factors interact with cis-regulatory elements to regulate gene expression. TRE mediates expression profiles of a large number of genes via trans-acting factors. While TRE mutations affect gene expression, it is also one of the main driving factors for evolutionary divergence in gene expression.

FourU thermometers are a class of non-coding RNA thermometers found in Salmonella. They are named 'FourU' due to the four highly conserved uridine nucleotides found directly opposite the Shine-Dalgarno sequence on hairpin II (pictured). RNA thermometers such as FourU control regulation of temperature via heat shock proteins in many prokaryotes. FourU thermometers are relatively small RNA molecules, only 57 nucleotides in length, and have a simple two-hairpin structure.
cspA mRNA 5' UTR is the untranslated region of the cspA gene, which is important in the cold shock response in Enterobacteriales such as E. coli. The 5' UTR element acts as an RNA thermometer, regulating the expression of cspA in response to temperature. By regulating temperature, cspA proteins carry out the vital function of homeostasis.

The developmentally active and heat shock inducible hsromega or hsrω gene in Drosophila produces multiple long non-coding RNA transcripts. This gene is transcriptionally active in almost all cell types of Drosophila and is the most actively induced following heat shock. A unique feature of the hsromega gene, which led to discovery of the 93D puff in 1970, is its singular inducibility with benzamide and a variety of other amides.

An RNA thermometer is a temperature-sensitive non-coding RNA molecule which regulates gene expression. RNA thermometers often regulate genes required during either a heat shock or cold shock response, but have been implicated in other regulatory roles such as in pathogenicity and starvation.
The IbpB thermometer is an RNA thermometer element found in the ibpAB operon. The operon contains two heat-shock genes, encoding inclusion body binding proteins A and B (IbpA/B), and is the most drastically upregulated operon under heat-shock in Escherichia coli.

In molecular biology, the Hsp17 thermometer is an RNA element found in the 5' UTR of Hsp17 mRNA. Hsp17 is a cyanobacterial heat shock protein belonging to the Hsp20 family.

The first cyanobacterial RNA thermometer (RNAT) Hsp17 was found in the 5'UTR of Synechocystis heat shock hsp17 mRNA. Further study demonstrated that cyanobacteria commonly use RNATs to control the translation of their heat shock genes. HspA is a homolog of Hsp17 in thermophilic Thermosynechococcus elongatus. Two more thermometers were found in the 5'UTRs of mesophilic cyanobacteria A. variabilis and Nostocsp. The first RNAT called avashort was shown to regulate translation by masking the AUG translation start site. The second RNAT called avalong, as it has an extended initial hairpin, might involve tertiary interactions and has similarities to the ROSE element.

Smaug is a RNA binding protein in Drosophila that helps in maternal to zygotic transition (MZT). The protein is named after the fictional character Smaug, the dragon in J.R.R. Tolkien's 1937 novel The Hobbit. The MZT ends with the midblastula transition (MBT), which is defined as the first developmental event in Drosophila that depends on zygotic mRNA. In Drosophila, the initial developmental events are controlled by maternal mRNAs like Hsp83, nanos, string, Pgc, and cyclin B mRNA. Degradation of these mRNAs, which is expected to terminate maternal control and enable zygotic control of embryogenesis, happens at interphase of nuclear division cycle 14. During this transition smaug protein targets the maternal mRNA for destruction using miRs. Thus activating the zygotic genes. Smaug is expected to play a role in expression of three miRNAs – miR-3, miR-6, miR-309 and miR-286 during MZT in Drosophila. Among them smaug dependent expression of miR-309 is needed for destabilization of 410 maternal mRNAs. In smaug mutants almost 85% of maternal mRNA is found to be stable. Smaug also binds to 3′ untranslated region (UTR) elements known as SMG response elements (SREs) on nanos mRNA and represses its expression by recruiting a protein called Cup(an eIF4E-binding protein that blocks the binding of eIF4G to eIF4E). There after it recruits deadenylation complex CCR4-Not on to the nanos mRNA which leads to deadenylation and subsequent decay of the mRNA. It is also found to be involved in degradation and repression of maternal Hsp83 mRNA by recruiting CCR4/POP2/NOT deadenylase to the mRNA. The human Smaug protein homologs are SAMD4A and SAMD4B.















