The start codon is the first codon of a messenger RNA (mRNA) transcript translated by a ribosome. The start codon always codes for methionine in eukaryotes and archaea and a N-formylmethionine (fMet) in bacteria, mitochondria and plastids.
Bacterial translation is the process by which messenger RNA is translated into proteins in bacteria.
In genetics, attenuation is a regulatory mechanism for some bacterial operons that results in premature termination of transcription. The canonical example of attenuation used in many introductory genetics textbooks, is ribosome-mediated attenuation of the trp operon. Ribosome-mediated attenuation of the trp operon relies on the fact that, in bacteria, transcription and translation proceed simultaneously. Attenuation involves a provisional stop signal (attenuator), located in the DNA segment that corresponds to the leader sequence of mRNA. During attenuation, the ribosome becomes stalled (delayed) in the attenuator region in the mRNA leader. Depending on the metabolic conditions, the attenuator either stops transcription at that point or allows read-through to the structural gene part of the mRNA and synthesis of the appropriate protein.
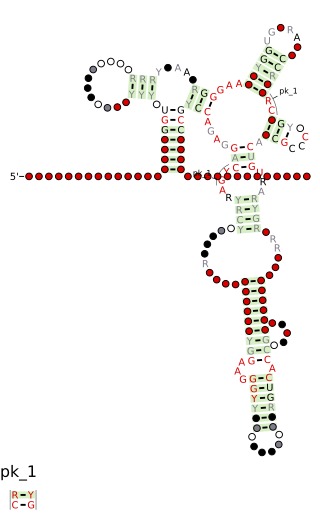
Cobalamin riboswitch is a cis-regulatory element which is widely distributed in 5' untranslated regions of vitamin B12 (Cobalamin) related genes in bacteria.
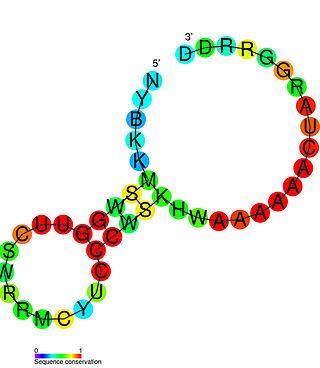
The PreQ1-I riboswitch is a cis-acting element identified in bacteria which regulates expression of genes involved in biosynthesis of the nucleoside queuosine (Q) from GTP. PreQ1 (pre-queuosine1) is an intermediate in the queuosine pathway, and preQ1 riboswitch, as a type of riboswitch, is an RNA element that binds preQ1. The preQ1 riboswitch is distinguished by its unusually small aptamer, compared to other riboswitches. Its atomic-resolution three-dimensional structure has been determined, with the PDB ID 2L1V.

A purine riboswitch is a sequence of ribonucleotides in certain messenger RNA (mRNA) that selectively binds to purine ligands via a natural aptamer domain. This binding causes a conformational change in the mRNA that can affect translation by revealing an expression platform for a downstream gene, or by forming a translation-terminating stem-loop. The ultimate effects of such translational regulation often take action to manage an abundance of the instigating purine, and might produce proteins that facilitate purine metabolism or purine membrane uptake.

The repression of heat shock gene expression (ROSE) element is an RNA element found in the 5' UTR of some heat shock protein's mRNAs. The ROSE element is an RNA thermometer that negatively regulates heat shock gene expression. The secondary structure is thought to be altered by temperature, thus it is an RNA thermometer. This structure blocks access to the ribosome binding site at normal temperatures. During heat shock however, the structure changes freeing the ribosome binding site and allowing expression to occur.
A ribosome binding site, or ribosomal binding site (RBS), is a sequence of nucleotides upstream of the start codon of an mRNA transcript that is responsible for the recruitment of a ribosome during the initiation of translation. Mostly, RBS refers to bacterial sequences, although internal ribosome entry sites (IRES) have been described in mRNAs of eukaryotic cells or viruses that infect eukaryotes. Ribosome recruitment in eukaryotes is generally mediated by the 5' cap present on eukaryotic mRNAs.

In molecular genetics, an untranslated region refers to either of two sections, one on each side of a coding sequence on a strand of mRNA. If it is found on the 5' side, it is called the 5' UTR, or if it is found on the 3' side, it is called the 3' UTR. mRNA is RNA that carries information from DNA to the ribosome, the site of protein synthesis (translation) within a cell. The mRNA is initially transcribed from the corresponding DNA sequence and then translated into protein. However, several regions of the mRNA are usually not translated into protein, including the 5' and 3' UTRs.
Cold shock response is a series of neurogenic cardio-respiratory responses caused by sudden immersion in cold water.

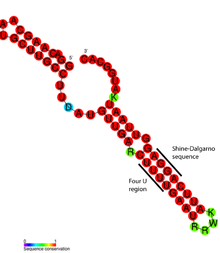
FourU thermometers are a class of non-coding RNA thermometers found in Salmonella. They are named 'FourU' due to the four highly conserved uridine nucleotides found directly opposite the Shine-Dalgarno sequence on hairpin II (pictured). RNA thermometers such as FourU control regulation of temperature via heat shock proteins in many prokaryotes. FourU thermometers are relatively small RNA molecules, only 57 nucleotides in length, and have a simple two-hairpin structure.

cspA mRNA 5' UTR is the untranslated region of the cspA gene, which is important in the cold shock response in Enterobacteriales such as E. coli. The 5' UTR element acts as an RNA thermometer, regulating the expression of cspA in response to temperature. By regulating temperature, cspA proteins carry out the vital function of homeostasis.
Bacterial small RNAs are small RNAs produced by bacteria; they are 50- to 500-nucleotide non-coding RNA molecules, highly structured and containing several stem-loops. Numerous sRNAs have been identified using both computational analysis and laboratory-based techniques such as Northern blotting, microarrays and RNA-Seq in a number of bacterial species including Escherichia coli, the model pathogen Salmonella, the nitrogen-fixing alphaproteobacterium Sinorhizobium meliloti, marine cyanobacteria, Francisella tularensis, Streptococcus pyogenes, the pathogen Staphylococcus aureus, and the plant pathogen Xanthomonas oryzae pathovar oryzae. Bacterial sRNAs affect how genes are expressed within bacterial cells via interaction with mRNA or protein, and thus can affect a variety of bacterial functions like metabolism, virulence, environmental stress response, and structure.

Escherichia coli contains a number of small RNAs located in intergenic regions of its genome. The presence of at least 55 of these has been verified experimentally. 275 potential sRNA-encoding loci were identified computationally using the QRNA program. These loci will include false positives, so the number of sRNA genes in E. coli is likely to be less than 275. A computational screen based on promoter sequences recognised by the sigma factor sigma 70 and on Rho-independent terminators predicted 24 putative sRNA genes, 14 of these were verified experimentally by northern blotting. The experimentally verified sRNAs included the well characterised sRNAs RprA and RyhB. Many of the sRNAs identified in this screen, including RprA, RyhB, SraB and SraL, are only expressed in the stationary phase of bacterial cell growth. A screen for sRNA genes based on homology to Salmonella and Klebsiella identified 59 candidate sRNA genes. From this set of candidate genes, microarray analysis and northern blotting confirmed the existence of 17 previously undescribed sRNAs, many of which bind to the chaperone protein Hfq and regulate the translation of RpoS. UptR sRNA transcribed from the uptR gene is implicated in suppressing extracytoplasmic toxicity by reducing the amount of membrane-bound toxic hybrid protein.

The IbpB thermometer is an RNA thermometer element found in the ibpAB operon. The operon contains two heat-shock genes, encoding inclusion body binding proteins A and B (IbpA/B), and is the most drastically upregulated operon under heat-shock in Escherichia coli.
In molecular biology, Toxic Small RNA(tsRNA, not to be confused with tRNA-derived small RNA) is a family of trans-encoded small non-coding RNA found exclusively in intergenic regions of Betaproteobacteria. Several paralogous loci may encode similar tsRNAs in each coding genome. Typically, each species of Burkholderia has 3-5 homologous tsRNAs. Experiments with four species of the Burkholderia lineage showed conserved and constitutive expression of tsRNAs in logarithmic growth phases.

In molecular biology, the Hsp17 thermometer is an RNA element found in the 5' UTR of Hsp17 mRNA. Hsp17 is a cyanobacterial heat shock protein belonging to the Hsp20 family.

The first cyanobacterial RNA thermometer (RNAT) Hsp17 was found in the 5'UTR of Synechocystis heat shock hsp17 mRNA. Further study demonstrated that cyanobacteria commonly use RNATs to control the translation of their heat shock genes. HspA is a homolog of Hsp17 in thermophilic Thermosynechococcus elongatus. Two more thermometers were found in the 5'UTRs of mesophilic cyanobacteria A. variabilis and Nostocsp. The first RNAT called avashort was shown to regulate translation by masking the AUG translation start site. The second RNAT called avalong, as it has an extended initial hairpin, might involve tertiary interactions and has similarities to the ROSE element.

RNA thermometers regulate gene expression in response to temperature allowing pathogens like Yersinia to switch on silent genes after entering the host organism. Usually, RNA thermometers are located in the 5'UTR, but an intergenic RNA thermometer was found in Yersinia pseudotuberculosis. The LcrFRNA thermometer together with the thermo-labile YmoA protein activates synthesis of the most crucial virulence activator LcrF (VirF). The RNA thermosensor sequence is 100% identical in all human pathogenic Yersinia species.
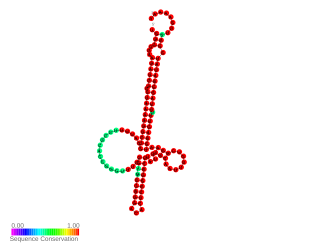
RNA thermometers (RNATs) regulate gene expression in response to temperature, allowing pathogens such as Neisseria meningitidis to switch on silent genes after entering the host organism. However the temperature for expression of Neisseria virulence-associated traits is 42 °C while other bacterial pathogen RNATs require 37 °C. This is probably because N. meningitidis is an obligate commensal of the human nasopharynx and becomes pathogenic during inflammation due to viral infection. Three independent RNA thermosensors were identified in the 5′UTRs of genes needed for: capsule biosynthesis (cssA), the expression of factor H binding protein (fHbp) and sialylation of lipopolysaccharide, which is essential for bacterial resistance against immune killing (lst). The very different nucleotide sequence and predicted inhibitory structures of the three RNATs indicate that they have evolved independently.