
In molecular biology, molecular chaperones are proteins that assist the conformational folding or unfolding of large proteins or macromolecular protein complexes. There are a number of classes of molecular chaperones, all of which function to assist large proteins in proper protein folding during or after synthesis, and after partial denaturation. Chaperones are also involved in the translocation of proteins for proteolysis.
Heat shock proteins (HSP) are a family of proteins produced by cells in response to exposure to stressful conditions. They were first described in relation to heat shock, but are now known to also be expressed during other stresses including exposure to cold, UV light and during wound healing or tissue remodeling. Many members of this group perform chaperone functions by stabilizing new proteins to ensure correct folding or by helping to refold proteins that were damaged by the cell stress. This increase in expression is transcriptionally regulated. The dramatic upregulation of the heat shock proteins is a key part of the heat shock response and is induced primarily by heat shock factor (HSF). HSPs are found in virtually all living organisms, from bacteria to humans.
In genetics, attenuation is a regulatory mechanism for some bacterial operons that results in premature termination of transcription. The canonical example of attenuation used in many introductory genetics textbooks, is ribosome-mediated attenuation of the trp operon. Ribosome-mediated attenuation of the trp operon relies on the fact that, in bacteria, transcription and translation proceed simultaneously. Attenuation involves a provisional stop signal (attenuator), located in the DNA segment that corresponds to the leader sequence of mRNA. During attenuation, the ribosome becomes stalled (delayed) in the attenuator region in the mRNA leader. Depending on the metabolic conditions, the attenuator either stops transcription at that point or allows read-through to the structural gene part of the mRNA and synthesis of the appropriate protein.

The trp operon is a group of genes that are transcribed together, encoding the enzymes that produce the amino acid tryptophan in bacteria. The trp operon was first characterized in Escherichia coli, and it has since been discovered in many other bacteria. The operon is regulated so that, when tryptophan is present in the environment, the genes for tryptophan synthesis are repressed.
The gene rpoS encodes the sigma factor sigma-38, a 37.8 kD protein in Escherichia coli. Sigma factors are proteins that regulate transcription in bacteria. Sigma factors can be activated in response to different environmental conditions. rpoS is transcribed in late exponential phase, and RpoS is the primary regulator of stationary phase genes. RpoS is a central regulator of the general stress response and operates in both a retroactive and a proactive manner: it not only allows the cell to survive environmental challenges, but it also prepares the cell for subsequent stresses (cross-protection). The transcriptional regulator CsgD is central to biofilm formation, controlling the expression of the curli structural and export proteins, and the diguanylate cyclase, adrA, which indirectly activates cellulose production. The rpoS gene most likely originated in the gammaproteobacteria.

fis is an E. coli gene encoding the Fis protein. The regulation of this gene is more complex than most other genes in the E. coli genome, as Fis is an important protein which regulates expression of other genes. It is supposed that fis is regulated by H-NS, IHF and CRP. It also regulates its own expression (autoregulation). Fis is one of the most abundant DNA binding proteins in Escherichia coli under nutrient-rich growth conditions.

The gcvB RNA gene encodes a small non-coding RNA involved in the regulation of a number of amino acid transport systems as well as amino acid biosynthetic genes. The GcvB gene is found in enteric bacteria such as Escherichia coli. GcvB regulates genes by acting as an antisense binding partner of the mRNAs for each regulated gene. This binding is dependent on binding to a protein called Hfq. Transcription of the GcvB RNA is activated by the adjacent GcvA gene and repressed by the GcvR gene. A deletion of GcvB RNA from Y. pestis changed colony shape as well as reducing growth. It has been shown by gene deletion that GcvB is a regulator of acid resistance in E. coli. GcvB enhances the ability of the bacterium to survive low pH by upregulating the levels of the alternate sigma factor RpoS. A polymeric form of GcvB has recently been identified. Interaction of GcvB with small RNA SroC triggers the degradation of GcvB by RNase E, lifting the GcvB-mediated mRNA repression of its target genes.

The repression of heat shock gene expression (ROSE) element is an RNA element found in the 5' UTR of some heat shock protein's mRNAs. The ROSE element is an RNA thermometer that negatively regulates heat shock gene expression. The secondary structure is thought to be altered by temperature, thus it is an RNA thermometer. This structure blocks access to the ribosome binding site at normal temperatures. During heat shock however, the structure changes freeing the ribosome binding site and allowing expression to occur.

Spot 42 (spf) RNA is a regulatory non-coding bacterial small RNA encoded by the spf gene. Spf is found in gammaproteobacteria and the majority of experimental work on Spot42 has been performed in Escherichia coli and recently in Aliivibrio salmonicida. In the cell Spot42 plays essential roles as a regulator in carbohydrate metabolism and uptake, and its expression is activated by glucose, and inhibited by the cAMP-CRP complex.
The GlmY RNA family consists of a number of bacterial RNA genes of around 167 bases in length. The GlmY RNA gene is present in Escherichia coli, Shigella flexneri, Yersinia pestis and Salmonella species, where it is found between the yfhK and purL genes. It was originally predicted in a bioinformatic screen for novel ncRNAs in E. coli.

The IscR stability element is a conserved secondary structure found in the intergenic regions of iscRSUA polycistronic mRNA. This secondary structure prevents the degradation of the iscR mRNA.

The JUMPstart RNA motif describes a conserved RNA-based secondary structure associated with JUMPstart elements. The 39-base-pair JUMPstart sequence describes a conserved element upstream of genes that participate in polysaccharide synthesis. The JUMPstart element has been shown to function as an RNA, and is present in the 5' untranslated regions of the genes it regulates.

FourU thermometers are a class of non-coding RNA thermometers found in Salmonella. They are named 'FourU' due to the four highly conserved uridine nucleotides found directly opposite the Shine-Dalgarno sequence on hairpin II (pictured). RNA thermometers such as FourU control regulation of temperature via heat shock proteins in many prokaryotes. FourU thermometers are relatively small RNA molecules, only 57 nucleotides in length, and have a simple two-hairpin structure.

cspA mRNA 5' UTR is the untranslated region of the cspA gene, which is important in the cold shock response in Enterobacteriales such as E. coli. The 5' UTR element acts as an RNA thermometer, regulating the expression of cspA in response to temperature. By regulating temperature, cspA proteins carry out the vital function of homeostasis.

Escherichia coli contains a number of small RNAs located in intergenic regions of its genome. The presence of at least 55 of these has been verified experimentally. 275 potential sRNA-encoding loci were identified computationally using the QRNA program. These loci will include false positives, so the number of sRNA genes in E. coli is likely to be less than 275. A computational screen based on promoter sequences recognised by the sigma factor sigma 70 and on Rho-independent terminators predicted 24 putative sRNA genes, 14 of these were verified experimentally by northern blotting. The experimentally verified sRNAs included the well characterised sRNAs RprA and RyhB. Many of the sRNAs identified in this screen, including RprA, RyhB, SraB and SraL, are only expressed in the stationary phase of bacterial cell growth. A screen for sRNA genes based on homology to Salmonella and Klebsiella identified 59 candidate sRNA genes. From this set of candidate genes, microarray analysis and northern blotting confirmed the existence of 17 previously undescribed sRNAs, many of which bind to the chaperone protein Hfq and regulate the translation of RpoS. UptR sRNA transcribed from the uptR gene is implicated in suppressing extracytoplasmic toxicity by reducing the amount of membrane-bound toxic hybrid protein.

An RNA thermometer is a temperature-sensitive non-coding RNA molecule which regulates gene expression. RNA thermometers often regulate genes required during either a heat shock or cold shock response, but have been implicated in other regulatory roles such as in pathogenicity and starvation.

In molecular biology, the Hsp17 thermometer is an RNA element found in the 5' UTR of Hsp17 mRNA. Hsp17 is a cyanobacterial heat shock protein belonging to the Hsp20 family.

The first cyanobacterial RNA thermometer (RNAT) Hsp17 was found in the 5'UTR of Synechocystis heat shock hsp17 mRNA. Further study demonstrated that cyanobacteria commonly use RNATs to control the translation of their heat shock genes. HspA is a homolog of Hsp17 in thermophilic Thermosynechococcus elongatus. Two more thermometers were found in the 5'UTRs of mesophilic cyanobacteria A. variabilis and Nostocsp. The first RNAT called avashort was shown to regulate translation by masking the AUG translation start site. The second RNAT called avalong, as it has an extended initial hairpin, might involve tertiary interactions and has similarities to the ROSE element.
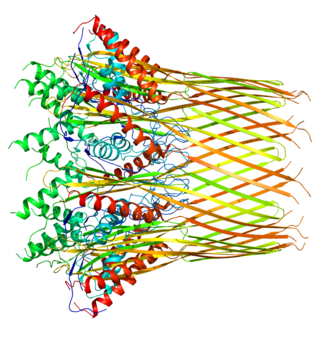
The Curli protein is a type of amyloid fiber produced by certain strains of enterobacteria. They are extracellular fibers located on bacteria such as E. coli and Salmonella spp. These fibers serve to promote cell community behavior through biofilm formation in the extracellular matrix. Amyloids are associated with several human neurodegenerative diseases such as Alzheimer's disease, Huntington's disease, Parkinson's disease, and prion diseases. The study of curli may help to understand human diseases thought to arise from improper amyloid fiber formation. The curli pili are generally assembled through the extracellular nucleation/precipitation pathway.
Catherine Louise Kearney Squires was a microbiologist known for her work on ribosomal RNA using Escherichia coli as a model organism. She was an elected fellow of the American Academy of Microbiology and the American Association for the Advancement of Science.















