
In the fields of molecular biology and genetics, a genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA. The nuclear genome includes protein-coding genes and non-coding genes, other functional regions of the genome such as regulatory sequences, and often a substantial fraction of junk DNA with no evident function. Almost all eukaryotes have mitochondria and a small mitochondrial genome. Algae and plants also contain chloroplasts with a chloroplast genome.

In biology, a mutation is an alteration in the nucleic acid sequence of the genome of an organism, virus, or extrachromosomal DNA. Viral genomes contain either DNA or RNA. Mutations result from errors during DNA or viral replication, mitosis, or meiosis or other types of damage to DNA, which then may undergo error-prone repair, cause an error during other forms of repair, or cause an error during replication. Mutations may also result from insertion or deletion of segments of DNA due to mobile genetic elements.

The human genome is a complete set of nucleic acid sequences for humans, encoded as DNA within the 23 chromosome pairs in cell nuclei and in a small DNA molecule found within individual mitochondria. These are usually treated separately as the nuclear genome and the mitochondrial genome. Human genomes include both protein-coding DNA sequences and various types of DNA that does not encode proteins. The latter is a diverse category that includes DNA coding for non-translated RNA, such as that for ribosomal RNA, transfer RNA, ribozymes, small nuclear RNAs, and several types of regulatory RNAs. It also includes promoters and their associated gene-regulatory elements, DNA playing structural and replicatory roles, such as scaffolding regions, telomeres, centromeres, and origins of replication, plus large numbers of transposable elements, inserted viral DNA, non-functional pseudogenes and simple, highly repetitive sequences. Introns make up a large percentage of non-coding DNA. Some of this non-coding DNA is non-functional junk DNA, such as pseudogenes, but there is no firm consensus on the total amount of junk DNA.
Non-coding DNA (ncDNA) sequences are components of an organism's DNA that do not encode protein sequences. Some non-coding DNA is transcribed into functional non-coding RNA molecules. Other functional regions of the non-coding DNA fraction include regulatory sequences that control gene expression; scaffold attachment regions; origins of DNA replication; centromeres; and telomeres. Some non-coding regions appear to be mostly nonfunctional, such as introns, pseudogenes, intergenic DNA, and fragments of transposons and viruses. Regions that are completely nonfunctional are called junk DNA.

A non-coding RNA (ncRNA) is a functional RNA molecule that is not translated into a protein. The DNA sequence from which a functional non-coding RNA is transcribed is often called an RNA gene. Abundant and functionally important types of non-coding RNAs include transfer RNAs (tRNAs) and ribosomal RNAs (rRNAs), as well as small RNAs such as microRNAs, siRNAs, piRNAs, snoRNAs, snRNAs, exRNAs, scaRNAs and the long ncRNAs such as Xist and HOTAIR.

Regulation of gene expression, or gene regulation, includes a wide range of mechanisms that are used by cells to increase or decrease the production of specific gene products. Sophisticated programs of gene expression are widely observed in biology, for example to trigger developmental pathways, respond to environmental stimuli, or adapt to new food sources. Virtually any step of gene expression can be modulated, from transcriptional initiation, to RNA processing, and to the post-translational modification of a protein. Often, one gene regulator controls another, and so on, in a gene regulatory network.
Repeated sequences are short or long patterns that occur in multiple copies throughout the genome. In many organisms, a significant fraction of the genomic DNA is repetitive, with over two-thirds of the sequence consisting of repetitive elements in humans. Some of these repeated sequences are necessary for maintaining important genome structures such as telomeres or centromeres.

The Chimpanzee Genome Project was an effort to determine the DNA sequence of the chimpanzee genome. Sequencing began in 2005 and by 2013 twenty-four individual chimpanzees had been sequenced. This project was folded into the Great Ape Genome Project.

In genetics, a silencer is a DNA sequence capable of binding transcription regulation factors, called repressors. DNA contains genes and provides the template to produce messenger RNA (mRNA). That mRNA is then translated into proteins. When a repressor protein binds to the silencer region of DNA, RNA polymerase is prevented from transcribing the DNA sequence into RNA. With transcription blocked, the translation of RNA into proteins is impossible. Thus, silencers prevent genes from being expressed as proteins.

In biology, the word gene has two meanings. The Mendelian gene is a basic unit of heredity. The molecular gene is a sequence of nucleotides in DNA that is transcribed to produce a functional RNA. There are two types of molecular genes: protein-coding genes and non-coding genes.

Human accelerated regions (HARs), first described in August 2006, are a set of 49 segments of the human genome that are conserved throughout vertebrate evolution but are strikingly different in humans. They are named according to their degree of difference between humans and chimpanzees. Found by scanning through genomic databases of multiple species, some of these highly mutated areas may contribute to human-specific traits. Others may represent loss of functional mutations, possibly due to the action of biased gene conversion rather than adaptive evolution.

NPAS3 or Neuronal PAS domain protein 3 is a brain-enriched transcription factor belonging to the bHLH-PAS superfamily of transcription factors, the members of which carry out diverse functions, including circadian oscillations, neurogenesis, toxin metabolism, hypoxia, and tracheal development. NPAS3 contains basic helix-loop-helix structural motif and PAS domain, like the other proteins in the superfamily.

Long non-coding RNAs are a type of RNA, generally defined as transcripts more than 200 nucleotides that are not translated into protein. This arbitrary limit distinguishes long ncRNAs from small non-coding RNAs, such as microRNAs (miRNAs), small interfering RNAs (siRNAs), Piwi-interacting RNAs (piRNAs), small nucleolar RNAs (snoRNAs), and other short RNAs. Given that some lncRNAs have been reported to have the potential to encode small proteins or micro-peptides, the latest definition of lncRNA is a class of RNA molecules of over 200 nucleotides that have no or limited coding capacity. Long intervening/intergenic noncoding RNAs (lincRNAs) are sequences of lncRNA which do not overlap protein-coding genes.

MEG3 is a maternally expressed, imprinted long non-coding RNA gene. At least 12 different isoforms of MEG3 are generated by alternative splicing. Expression of MEG3 is lost in cancer cells. It acts as a growth suppressor in tumour cells, and activates p53. A pituitary transcript variant has been associated with inhibited cell proliferation. Studies in mouse and sheep suggest that an upstream intergenic differentially methylated region (IG-DMR) regulates imprinting of the region. The expression profile in mouse of the co-regulated Meg3 and Dlk1 genes suggests a causative role in the pathologies found in uniparental disomy animals, characterized by defects in skeletal muscle maturation, bone formation, placenta size and organization and prenatal lethality. The sheep homolog is associated with the callipyge mutation which in heterozygous individuals affects a muscle-specific long-range control element located in the DLK1-GTL2 intergenic region and results in the callipyge muscular hypertrophy. The non-Mendelian inheritance pattern, known as polar overdominance, likely results from the combination of the cis-effect on the expression levels of genes in the DLK1-GTL2 imprinted domain, and trans interaction between the products of reciprocally imprinted genes. MEG3 is thought to play a role in the development of Alzheimer's disease by triggering necroptosis.
A conserved non-coding sequence (CNS) is a DNA sequence of noncoding DNA that is evolutionarily conserved. These sequences are of interest for their potential to regulate gene production.

MIAT, also known as RNCR2 or Gomafu, is a long non-coding RNA. Single nucleotide polymorphisms (SNPs) in MIAT are associated with a risk of myocardial infarction. It is expressed in neurons, and located in the nucleus. It plays a role in the regulation of retinal cell fate specification. Crea and collaborators have shown that MIAT is highly up-regulated in aggressive prostate cancer samples, raising the possibility that this gene plays a role in cancer progression.
Genome instability refers to a high frequency of mutations within the genome of a cellular lineage. These mutations can include changes in nucleic acid sequences, chromosomal rearrangements or aneuploidy. Genome instability does occur in bacteria. In multicellular organisms genome instability is central to carcinogenesis, and in humans it is also a factor in some neurodegenerative diseases such as amyotrophic lateral sclerosis or the neuromuscular disease myotonic dystrophy.
Epigenetics of human development is the study of how epigenetics effects human development.
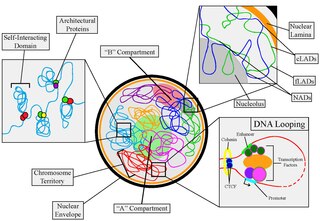
Nuclear organization refers to the spatial distribution of chromatin within a cell nucleus. There are many different levels and scales of nuclear organisation. Chromatin is a higher order structure of DNA.
Katherine Snowden Pollard is the Director of the Gladstone Institute of Data Science and Biotechnology and a professor at the University of California, San Francisco (UCSF). She is a Chan Zuckerberg Biohub Investigator. She was awarded Fellowship of the International Society for Computational Biology in 2020 and the American Institute for Medical and Biological Engineering in 2021 for outstanding contributions to computational biology and bioinformatics.












