Related Research Articles
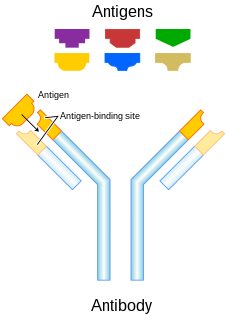
An antibody (Ab), also known as an immunoglobulin (Ig), is a large, Y-shaped protein used by the immune system to identify and neutralize foreign objects such as pathogenic bacteria and viruses. The antibody recognizes a unique molecule of the pathogen, called an antigen. Each tip of the "Y" of an antibody contains a paratope that is specific for one particular epitope on an antigen, allowing these two structures to bind together with precision. Using this binding mechanism, an antibody can tag a microbe or an infected cell for attack by other parts of the immune system, or can neutralize it directly.
In biology and biochemistry, protease inhibitors, or antiproteases, are molecules that inhibit the function of proteases. Many naturally occurring protease inhibitors are proteins.

Superantigens (SAgs) are a class of antigens that result in excessive activation of the immune system. Specifically it causes non-specific activation of T-cells resulting in polyclonal T cell activation and massive cytokine release. SAgs are produced by some pathogenic viruses and bacteria most likely as a defense mechanism against the immune system. Compared to a normal antigen-induced T-cell response where 0.0001-0.001% of the body's T-cells are activated, these SAgs are capable of activating up to 20% of the body's T-cells. Furthermore, Anti-CD3 and Anti-CD28 antibodies (CD28-SuperMAB) have also shown to be highly potent superantigens.

In molecular biology, intercellular adhesion molecules (ICAMs) and vascular cell adhesion molecule-1 (VCAM-1) are part of the immunoglobulin superfamily. They are important in inflammation, immune responses and in intracellular signalling events. The ICAM family consists of five members, designated ICAM-1 to ICAM-5. They are known to bind to leucocyte integrins CD11/CD18 such as LFA-1 and Macrophage-1 antigen, during inflammation and in immune responses. In addition, ICAMs may exist in soluble forms in human plasma, due to activation and proteolysis mechanisms at cell surfaces.

Laminins are high-molecular weight proteins of the extracellular matrix. They are a major component of the basal lamina, a protein network foundation for most cells and organs. The laminins are an important and biologically active part of the basal lamina, influencing cell differentiation, migration, and adhesion.

The immunoglobulin domain, also known as the immunoglobulin fold, is a type of protein domain that consists of a 2-layer sandwich of 7-9 antiparallel β-strands arranged in two β-sheets with a Greek key topology, consisting of about 125 amino acids.

Vascular cell adhesion protein 1 also known as vascular cell adhesion molecule 1 (VCAM-1) or cluster of differentiation 106 (CD106) is a protein that in humans is encoded by the VCAM1 gene. VCAM-1 functions as a cell adhesion molecule.

The immunoglobulin superfamily (IgSF) is a large protein superfamily of cell surface and soluble proteins that are involved in the recognition, binding, or adhesion processes of cells. Molecules are categorized as members of this superfamily based on shared structural features with immunoglobulins ; they all possess a domain known as an immunoglobulin domain or fold. Members of the IgSF include cell surface antigen receptors, co-receptors and co-stimulatory molecules of the immune system, molecules involved in antigen presentation to lymphocytes, cell adhesion molecules, certain cytokine receptors and intracellular muscle proteins. They are commonly associated with roles in the immune system. Otherwise, the sperm-specific protein IZUMO1, a member of the immunoglobulin superfamily, has also been identified as the only sperm membrane protein essential for sperm-egg fusion.

Complementarity-determining regions (CDRs) are part of the variable chains in immunoglobulins (antibodies) and T cell receptors, generated by B-cells and T-cells respectively, where these molecules bind to their specific antigen. A set of CDRs constitutes a paratope. As the most variable parts of the molecules, CDRs are crucial to the diversity of antigen specificities generated by lymphocytes.

Sialoadhesin is a cell adhesion molecule found on the surface of macrophages. It is found in especially high amounts on macrophages of the spleen, liver, lymph node, bone marrow, colon, and lungs. Also, in patients suffering from rheumatoid arthritis, the protein has been found in great amounts on macrophages of the affected tissues. It is defined as an I-type lectin, since it contains 17 immunoglobulin (Ig) domains, and thus also belongs to the immunoglobulin superfamily (IgSF). Sialoadhesin binds to certain molecules called sialic acids. During this binding process a salt bridge (protein) is formed between a highly conserved arginine residue and the carboxylate group of the sialic acid. Since sialoadhesin binds sialic acids with its N-terminal IgV-domain, it is also a member of the SIGLEC family. Alternate names for sialoadhesin include siglec-1 and CD169.

A protein domain is a region of the protein's polypeptide chain that is self-stabilizing and that folds independently from the rest. Each domain forms a compact folded three-dimensional structure. Many proteins consist of several domains. One domain may appear in a variety of different proteins. Molecular evolution uses domains as building blocks and these may be recombined in different arrangements to create proteins with different functions. In general, domains vary in length from between about 50 amino acids up to 250 amino acids in length. The shortest domains, such as zinc fingers, are stabilized by metal ions or disulfide bridges. Domains often form functional units, such as the calcium-binding EF hand domain of calmodulin. Because they are independently stable, domains can be "swapped" by genetic engineering between one protein and another to make chimeric proteins.
Immunoglobulin kappa constant, also known as IGKC, is a human gene that encodes the constant domain of kappa-type light chains for antibodies. It is found on chromosome 2, in humans, within the Immunoglobulin kappa locus, IGK@.

Neuronal cell adhesion molecule is a protein that in humans is encoded by the NRCAM gene.
Immunoglobulin heavy constant alpha 1 is a immunoglobulin gene with symbol IGHA1. It encodes a constant (C) segment of Immunoglobulin A heavy chain. Immunoglobulin A is an antibody that plays a critical role in immune function in the mucous membranes. IgA shows the same typical structure of other antibody classes, with two heavy chains and two light chains, and four distinct domains: one variable region, and three variable regions. As a major class of immunoglobulin in body secretions, IgA plays a role in defending against infection, as well as preventing the access of foreign antigens to the immunologic system.

Protocadherin gamma-C3 is a protein that in humans is encoded by the PCDHGC3 gene.

CD79b molecule, immunoglobulin-associated beta, also known as CD79B, is a human gene.
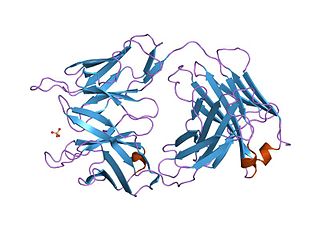
V-set domains are Ig-like domains resembling the antibody variable domain. V-set domains are found in diverse protein families, including immunoglobulin light and heavy chains; in several T-cell receptors such as CD2, CD4, CD80, and CD86; in myelin membrane adhesion molecules; in junctional adhesion molecules (JAM); in tyrosine-protein kinase receptors; and in the programmed cell death protein 1 (PD1).
IgSF CAMs are cell adhesion molecules that belong to Immunoglobulin superfamily. It is regarded as the most diverse superfamily of CAMs. This family is characterized by their extracellular domains containing Ig-like domains. The Ig domains are then followed by Fibronectin type III domain repeats and IgSFs are anchored to the membrane by a GPI moiety. This family is involved in both homophilic or heterophilic binding and has the ability to bind integrins or different IgSF CAMs.

In the field of molecular biology, enterotoxin type B, also known as Staphylococcal enterotoxin B (SEB), is an enterotoxin produced by the gram-positive bacteria Staphylococcus aureus. It is a common cause of food poisoning, with severe diarrhea, nausea and intestinal cramping often starting within a few hours of ingestion. Being quite stable, the toxin may remain active even after the contaminating bacteria are killed. It can withstand boiling at 100 °C for a few minutes. Gastroenteritis occurs because SEB is a superantigen, causing the immune system to release a large amount of cytokines that lead to significant inflammation.

In molecular biology, the adhesin molecule (immunoglobulin-like) is a protein domain. This domain is found in mucosal vascular addressin cell adhesion molecule 1 proteins (MAdCAM-1). These are cell adhesion molecules expressed on the endothelium in mucosa that guide the specific homing of lymphocytes into mucosal tissues. MAdCAM-1 belongs to a subclass of the immunoglobulin superfamily (IgSF), the members of which are ligands for integrins. The crystal structure of this domain has been reported; it adopts an immunoglobulin-like beta-sandwich structure, with seven strands arranged in two beta-sheets in a Greek-key topology.
References
- ↑ Smith DK, Xue H (1997). "Sequence profiles of immunoglobulin and immunoglobulin-like domains". J. Mol. Biol. 274 (4): 530–545. doi:10.1006/jmbi.1997.1432. PMID 9417933.
- ↑ Potapov V, Sobolev V, Edelman M, Kister A, Gelfand I (2004). "Protein-Protein Recognition: Juxtaposition of Domain and Interface Cores in Immunoglobulins and Other Sandwich-like Proteins". J. Mol. Biol. 342 (2): 665–679. doi:10.1016/j.jmb.2004.06.072. PMID 15327963.
- ↑ Clarke J, Fowler SB (2001). "Mapping the folding pathway of an immunoglobulin domain: structural detail from Phi value analysis and movement of the transition state". Structure. 9 (5): 355–366. doi: 10.1016/S0969-2126(01)00596-2 . PMID 11377196.
- ↑ Chothia C, Teichmann SA (2000). "Immunoglobulin superfamily proteins in Caenorhabditis elegans". J. Mol. Biol. 296 (5): 1367–83. CiteSeerX 10.1.1.327.6917 . doi:10.1006/jmbi.1999.3497. PMID 10698639.
- ↑ Reinherz EL, Yang H (2001). "Dynamic recruitment of human CD2 into lipid rafts. Linkage to T cell signal transduction". J. Biol. Chem. 276 (22): 18775–18785. doi: 10.1074/jbc.M009852200 . PMID 11376005.
- ↑ Kutish GF, Rock DL, Afonso CL, Borca MV, Irusta P, Carrillo C, Brun A, Sussman M (1994). "An African swine fever virus gene with similarity to the T-lymphocyte surface antigen CD2 mediates hemadsorption". Virology. 199 (2): 463–468. doi:10.1006/viro.1994.1146. PMID 7907198.
- ↑ Sanejouand YH (2004). "Domain swapping of CD4 upon dimerization". Proteins. 57 (1): 205–12. doi:10.1002/prot.20197. PMID 15326605.