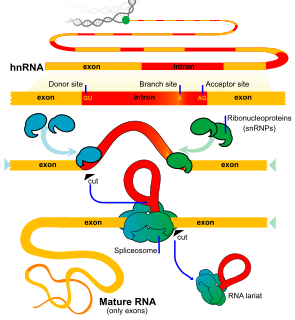
An exon is any part of a gene that will form a part of the final mature RNA produced by that gene after introns have been removed by RNA splicing. The term exon refers to both the DNA sequence within a gene and to the corresponding sequence in RNA transcripts. In RNA splicing, introns are removed and exons are covalently joined to one another as part of generating the mature RNA. Just as the entire set of genes for a species constitutes the genome, the entire set of exons constitutes the exome.
An intron is any nucleotide sequence within a gene that is removed by RNA processing during production of the final RNA product. The word intron is derived from the term intragenic region, i.e. a region inside a gene. The term intron refers to both the DNA sequence within a gene and the corresponding RNA sequence in RNA transcripts. The non-intron sequences that become joined by this RNA processing to form the mature RNA are called exons.

RNA splicing is a process in molecular biology where a newly-made precursor messenger RNA (pre-mRNA) transcript is transformed into a mature messenger RNA (mRNA). It works by removing all the introns and splicing back together exons. For nuclear-encoded genes, splicing occurs in the nucleus either during or immediately after transcription. For those eukaryotic genes that contain introns, splicing is usually needed to create an mRNA molecule that can be translated into protein. For many eukaryotic introns, splicing occurs in a series of reactions which are catalyzed by the spliceosome, a complex of small nuclear ribonucleoproteins (snRNPs). There exist self-splicing introns, that is, ribozymes that can catalyze their own excision from their parent RNA molecule. The process of transcription, splicing and translation is called gene expression, the central dogma of molecular biology.

Caenorhabditis elegans is a free-living transparent nematode about 1 mm in length that lives in temperate soil environments. It is the type species of its genus. The name is a blend of the Greek caeno- (recent), rhabditis (rod-like) and Latin elegans (elegant). In 1900, Maupas initially named it Rhabditides elegans. Osche placed it in the subgenus Caenorhabditis in 1952, and in 1955, Dougherty raised Caenorhabditis to the status of genus.

Alternative splicing, or alternative RNA splicing, or differential splicing, is an alternative splicing process during gene expression that allows a single gene to code for multiple proteins. In this process, particular exons of a gene may be included within or excluded from the final, processed messenger RNA (mRNA) produced from that gene. This means the exons are joined in different combinations, leading to different (alternative) mRNA strands. Consequently, the proteins translated from alternatively spliced mRNAs will contain differences in their amino acid sequence and, often, in their biological functions.
Trans-splicing is a special form of RNA processing where exons from two different primary RNA transcripts are joined end to end and ligated. It is usually found in eukaryotes and mediated by the spliceosome, although some bacteria and archaea also have "half-genes" for tRNAs.

Mitochondrial 5-demethoxyubiquinone hydroxylase, also known as coenzyme Q7, hydroxylase, is an enzyme that in humans is encoded by the COQ7 gene. The clk-1 (clock-1) gene encodes this protein that is necessary for ubiquinone biosynthesis in the worm Caenorhabditis elegans and other eukaryotes. The mouse version of the gene is called mclk-1 and the human, fruit fly and yeast homolog COQ7.

Caenorhabditis is a genus of nematodes which live in bacteria-rich environments like compost piles, decaying dead animals and rotting fruit. The name comes from Greek: caeno- ; rhabditis = rod-like. In 1900, Maupas initially named the species Rhabditis elegans, Osche placed it in the subgenus Caenorhabditis in 1952, and in 1955, Dougherty raised Caenorhabditis to the status of genus.

U6 snRNA is the non-coding small nuclear RNA (snRNA) component of U6 snRNP, an RNA-protein complex that combines with other snRNPs, unmodified pre-mRNA, and various other proteins to assemble a spliceosome, a large RNA-protein molecular complex that catalyzes the excision of introns from pre-mRNA. Splicing, or the removal of introns, is a major aspect of post-transcriptional modification and takes place only in the nucleus of eukaryotes.
RNA binding motif protein 9 (RBM9), also known as Rbfox2, is a protein which in humans is encoded by the RBM9 gene.
Fox-1 homolog A, also known as ataxin 2-binding protein 1 (A2BP1) or hexaribonucleotide-binding protein 1 (HRNBP1) or RNA binding protein, fox-1 homolog (Rbfox1), is a protein that in humans is encoded by the RBFOX1 gene.
Trans-Spliced Exon Coupled RNA End Determination (TEC-RED) is a transcriptomic technique that, like SAGE, allows for the digital detection of messenger RNA sequences. Unlike SAGE, detection and purification of transcripts from the 5’ end of the messenger RNA require the presence of a trans-spliced leader sequence.

28S ribosomal protein S33, mitochondrial is a protein that in humans is encoded by the MRPS33 gene.
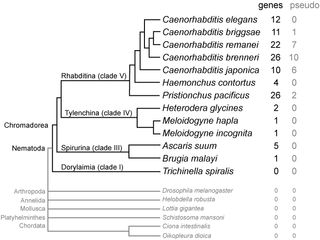
SmY ribonucleic acids are a family of small nuclear RNAs found in some species of nematode worms. They are thought to be involved in mRNA trans-splicing.
Mirtrons are a type of microRNAs that are located in the introns of the mRNA encoding host genes. These short hairpin introns formed via atypical miRNA biogenesis pathways. Mirtrons arise from the spliced-out introns and are known to function in gene expression.
Periannan Senapathy is a molecular biologist, geneticist, author and entrepreneur. He is the founder, president and chief scientific officer at Genome International Corporation, a biotechnology, bioinformatics, and information technology firm based in Madison, Wisconsin, which develops computational genomics applications of next-generation DNA sequencing (NGS) and clinical decision support systems for analyzing patient genome data that aids in diagnosis and treatment of diseases.
High-throughput sequencing of RNA isolated by crosslinking immunoprecipitation is a genome-wide means of mapping protein–RNA binding sites or RNA modification sites in vivo. HITS-CLIP was originally used to generate genome-wide protein-RNA interaction maps for the neuron-specific RNA-binding protein and splicing factor NOVA1 and NOVA2; since then a number of other splicing factor maps have been generated, including those for PTB, RbFox2, SFRS1, hnRNP C, and even N6-Methyladenosine (m6A) mRNA modifications.

NeuN , a protein which is a homologue to the protein product of a sex-determining gene in Caenorhabditis elegans, is a neuronal nuclear antigen that is commonly used as a biomarker for neurons.
WormBase is an online biological database about the biology and genome of the nematode model organism Caenorhabditis elegans and contains information about other related nematodes. WormBase is used by the C. elegans research community both as an information resource and as a place to publish and distribute their results. The database is regularly updated with new versions being released every two months. WormBase is one of the organizations participating in the Generic Model Organism Database (GMOD) project.
An outron is a nucleotide sequence at the 5' end of the primary transcript of a gene that is removed by a special form of RNA splicing during maturation of the final RNA product. Whereas intron sequences are located inside the gene, outron sequences lie outside the gene.










