
Evolutionary developmental biology is a field of biological research that compares the developmental processes of different organisms to infer how developmental processes evolved.

A non-coding RNA (ncRNA) is a functional RNA molecule that is not translated into a protein. The DNA sequence from which a functional non-coding RNA is transcribed is often called an RNA gene. Abundant and functionally important types of non-coding RNAs include transfer RNAs (tRNAs) and ribosomal RNAs (rRNAs), as well as small RNAs such as microRNAs, siRNAs, piRNAs, snoRNAs, snRNAs, exRNAs, scaRNAs and the long ncRNAs such as Xist and HOTAIR.

A generegulatory network (GRN) is a collection of molecular regulators that interact with each other and with other substances in the cell to govern the gene expression levels of mRNA and proteins which, in turn, determine the function of the cell. GRN also play a central role in morphogenesis, the creation of body structures, which in turn is central to evolutionary developmental biology (evo-devo).

Eugene Viktorovich Koonin is a Russian-American biologist and Senior Investigator at the National Center for Biotechnology Information (NCBI). He is a recognised expert in the field of evolutionary and computational biology.

Comparative genomics is a branch of biological research that examines genome sequences across a spectrum of species, spanning from humans and mice to a diverse array of organisms from bacteria to chimpanzees. This large-scale holistic approach compares two or more genomes to discover the similarities and differences between the genomes and to study the biology of the individual genomes. Comparison of whole genome sequences provides a highly detailed view of how organisms are related to each other at the gene level. By comparing whole genome sequences, researchers gain insights into genetic relationships between organisms and study evolutionary changes. The major principle of comparative genomics is that common features of two organisms will often be encoded within the DNA that is evolutionarily conserved between them. Therefore, Comparative genomics provides a powerful tool for studying evolutionary changes among organisms, helping to identify genes that are conserved or common among species, as well as genes that give unique characteristics of each organism. Moreover, these studies can be performed at different levels of the genomes to obtain multiple perspectives about the organisms.
The transcriptome is the set of all RNA transcripts, including coding and non-coding, in an individual or a population of cells. The term can also sometimes be used to refer to all RNAs, or just mRNA, depending on the particular experiment. The term transcriptome is a portmanteau of the words transcript and genome; it is associated with the process of transcript production during the biological process of transcription.

In evolutionary biology, conserved sequences are identical or similar sequences in nucleic acids or proteins across species, or within a genome, or between donor and receptor taxa. Conservation indicates that a sequence has been maintained by natural selection.
Ectopic is a word used with a prefix ecto-, meaning “out of”, and the suffix -topic, meaning "place." Ectopic expression is an abnormal gene expression in a cell type, tissue type, or developmental stage in which the gene is not usually expressed. The term ectopic expression is predominantly used in studies using metazoans, especially in Drosophila melanogaster for research purposes.

The Argonaute protein family, first discovered for its evolutionarily conserved stem cell function, plays a central role in RNA silencing processes as essential components of the RNA-induced silencing complex (RISC). RISC is responsible for the gene silencing phenomenon known as RNA interference (RNAi). Argonaute proteins bind different classes of small non-coding RNAs, including microRNAs (miRNAs), small interfering RNAs (siRNAs) and Piwi-interacting RNAs (piRNAs). Small RNAs guide Argonaute proteins to their specific targets through sequence complementarity, which then leads to mRNA cleavage, translation inhibition, and/or the initiation of mRNA decay.

Victor R. Ambros is an American developmental biologist who discovered the first known microRNA (miRNA). He is a professor at the University of Massachusetts Medical School in Worcester, Massachusetts.
Gary Bruce Ruvkun is an American molecular biologist at Massachusetts General Hospital and professor of genetics at Harvard Medical School in Boston. Ruvkun discovered the mechanism by which lin-4, the first microRNA (miRNA) discovered by Victor Ambros, regulates the translation of target messenger RNAs via imperfect base-pairing to those targets, and discovered the second miRNA, let-7, and that it is conserved across animal phylogeny, including in humans. These miRNA discoveries revealed a new world of RNA regulation at an unprecedented small size scale, and the mechanism of that regulation. Ruvkun also discovered many features of insulin-like signaling in the regulation of aging and metabolism. He was elected a Member of the American Philosophical Society in 2019.

RNA-Seq is a technique that uses next-generation sequencing to reveal the presence and quantity of RNA molecules in a biological sample, providing a snapshot of gene expression in the sample, also known as transcriptome.

Laurence Daniel Hurst is a Professor of Evolutionary Genetics in the Department of Biology and Biochemistry at the University of Bath and the director of the Milner Centre for Evolution.
Single-cell sequencing examines the nucleic acid sequence information from individual cells with optimized next-generation sequencing technologies, providing a higher resolution of cellular differences and a better understanding of the function of an individual cell in the context of its microenvironment. For example, in cancer, sequencing the DNA of individual cells can give information about mutations carried by small populations of cells. In development, sequencing the RNAs expressed by individual cells can give insight into the existence and behavior of different cell types. In microbial systems, a population of the same species can appear genetically clonal. Still, single-cell sequencing of RNA or epigenetic modifications can reveal cell-to-cell variability that may help populations rapidly adapt to survive in changing environments.
Claude Desplan, a biologist originally trained in France, has been a Silver Professor in New York University’s Department of Biology since 1999. His research centers on understanding the development and functioning of the visual system that underlies color vision using the fruit fly Drosophila as a model organism.

Sarah Amalia Teichmann is a German scientist who is head of cellular genetics at the Wellcome Sanger Institute and a visiting research group leader at the European Bioinformatics Institute (EMBL-EBI). She serves as director of research in the Cavendish Laboratory, at the University of Cambridge and a senior research fellow at Churchill College, Cambridge.
Scott Frederick Gilbert is an American evolutionary developmental biologist and historian of biology.
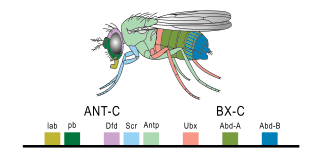
The evo-devo gene toolkit is the small subset of genes in an organism's genome whose products control the organism's embryonic development. Toolkit genes are central to the synthesis of molecular genetics, palaeontology, evolution and developmental biology in the science of evolutionary developmental biology (evo-devo). Many of them are ancient and highly conserved among animal phyla.
Transcriptomics technologies are the techniques used to study an organism's transcriptome, the sum of all of its RNA transcripts. The information content of an organism is recorded in the DNA of its genome and expressed through transcription. Here, mRNA serves as a transient intermediary molecule in the information network, whilst non-coding RNAs perform additional diverse functions. A transcriptome captures a snapshot in time of the total transcripts present in a cell. Transcriptomics technologies provide a broad account of which cellular processes are active and which are dormant. A major challenge in molecular biology is to understand how a single genome gives rise to a variety of cells. Another is how gene expression is regulated.

Micropeptides are polypeptides with a length of less than 100-150 amino acids that are encoded by short open reading frames (sORFs). In this respect, they differ from many other active small polypeptides, which are produced through the posttranslational cleavage of larger polypeptides. In terms of size, micropeptides are considerably shorter than "canonical" proteins, which have an average length of 330 and 449 amino acids in prokaryotes and eukaryotes, respectively. Micropeptides are sometimes named according to their genomic location. For example, the translated product of an upstream open reading frame (uORF) might be called a uORF-encoded peptide (uPEP). Micropeptides lack an N-terminal signaling sequences, suggesting that they are likely to be localized to the cytoplasm. However, some micropeptides have been found in other cell compartments, as indicated by the existence of transmembrane micropeptides. They are found in both prokaryotes and eukaryotes. The sORFs from which micropeptides are translated can be encoded in 5' UTRs, small genes, or polycistronic mRNAs. Some micropeptide-coding genes were originally mis-annotated as long non-coding RNAs (lncRNAs).












