
The trp operon is a group of genes that are transcribed together, encoding the enzymes that produce the amino acid tryptophan in bacteria. The trp operon was first characterized in Escherichia coli, and it has since been discovered in many other bacteria. The operon is regulated so that, when tryptophan is present in the environment, the genes for tryptophan synthesis are repressed.
cAMP receptor protein is a regulatory protein in bacteria. CRP protein binds cAMP, which causes a conformational change that allows CRP to bind tightly to a specific DNA site in the promoters of the genes it controls. CRP then activates transcription through direct protein–protein interactions with RNA polymerase.

fis is an E. coli gene encoding the Fis protein. The regulation of this gene is more complex than most other genes in the E. coli genome, as Fis is an important protein which regulates expression of other genes. It is supposed that fis is regulated by H-NS, IHF and CRP. It also regulates its own expression (autoregulation). Fis is one of the most abundant DNA binding proteins in Escherichia coli under nutrient-rich growth conditions.

The gcvB RNA gene encodes a small non-coding RNA involved in the regulation of a number of amino acid transport systems as well as amino acid biosynthetic genes. The GcvB gene is found in enteric bacteria such as Escherichia coli. GcvB regulates genes by acting as an antisense binding partner of the mRNAs for each regulated gene. This binding is dependent on binding to a protein called Hfq. Transcription of the GcvB RNA is activated by the adjacent GcvA gene and repressed by the GcvR gene. A deletion of GcvB RNA from Y. pestis changed colony shape as well as reducing growth. It has been shown by gene deletion that GcvB is a regulator of acid resistance in E. coli. GcvB enhances the ability of the bacterium to survive low pH by upregulating the levels of the alternate sigma factor RpoS. A polymeric form of GcvB has recently been identified. Interaction of GcvB with small RNA SroC triggers the degradation of GcvB by RNase E, lifting the GcvB-mediated mRNA repression of its target genes.
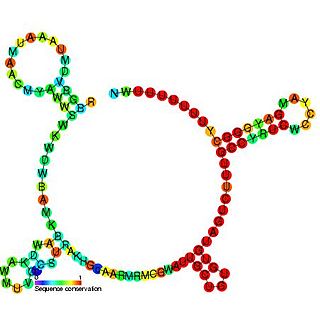
The RprA RNA gene encodes a 106 nucleotide regulatory non-coding RNA. Translational regulation of the stationary phase sigma factor RpoS is mediated by the formation of a double-stranded RNA stem-loop structure in the upstream region of the rpoS messenger RNA, occluding the translation initiation site.

The hok/sok system is a postsegregational killing mechanism employed by the R1 plasmid in Escherichia coli. It was the first type I toxin-antitoxin pair to be identified through characterisation of a plasmid-stabilising locus. It is a type I system because the toxin is neutralised by a complementary RNA, rather than a partnered protein.
In biology, phase variation is a method for dealing with rapidly varying environments without requiring random mutation. It involves the variation of protein expression, frequently in an on-off fashion, within different parts of a bacterial population. As such the phenotype can switch at frequencies that are much higher than classical mutation rates. Phase variation contributes to virulence by generating heterogeneity. Although it has been most commonly studied in the context of immune evasion, it is observed in many other areas as well and is employed by various types of bacteria, including Salmonella species.
The gal operon is a prokaryotic operon, which encodes enzymes necessary for galactose metabolism. Repression of gene expression for this operon works via binding of repressor molecules to two operators. These repressors dimerize, creating a loop in the DNA. The loop as well as hindrance from the external operator prevent RNA polymerase from binding to the promoter, and thus prevent transcription. Additionally, since the metabolism of galactose in the cell is involved in both anabolic and catabolic pathways, a novel regulatory system using two promoters for differential repression has been identified and characterized within the context of the gal operon.
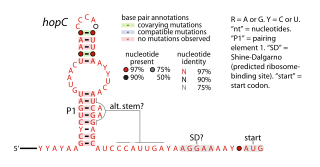
The hopC RNA motif is a predicted cis-regulatory element identified by a bioinformatic screen for conserved RNA secondary structures. hopC RNAs are exclusively found within bacteria classified within the genus Helicobacter, some of which are human pathogens that infect the stomach and can cause ulcers.

The psaA RNA motif describes a class of RNAs with a common secondary structure. psaA RNAs are exclusively found in locations that presumably correspond to the 5' untranslated regions of operons formed of psaA and psaB genes. For this reason, it was hypothesized that psaA RNAs function as cis-regulatory elements of these genes. The psaAB genes encode proteins that form subunits in the photosystem I structure used for photosynthesis. psaA RNAs have been detected only in cyanobacteria, which is consistent with their association with photosynthesis.

The rsmX gene is part of the Rsm/Csr family of non-coding RNAs (ncRNAs). Members of the Rsm/Csr family are present in a diverse range of bacteria, including Escherichia coli, Erwinia, Salmonella, Vibrio and Pseudomonas. These ncRNAs act by sequestering translational repressor proteins, called RsmA, activating expression of downstream genes that would normally be blocked by the repressors. Sequestering of target proteins is dependent upon exposed GGA motifs in the stem loops of the ncRNAs. Typically, the activated genes are involved in secondary metabolism, biofilm formation and motility.

The eps-Associated RNA element is a conserved RNA motif associated with exopolysaccharide (eps) or capsule biosynthesis genes in a subset of bacteria classified within the order Bacillales. It was initially discovered in Bacillus subtilis, located between the second and third gene in the eps operon. Deletion of the EAR element impairs biofilm formation.

cspA mRNA 5' UTR is the untranslated region of the cspA gene, which is important in the cold shock response in Enterobacteriales such as E. coli. The 5' UTR element acts as an RNA thermometer, regulating the expression of cspA in response to temperature. By regulating temperature, cspA proteins carry out the vital function of homeostasis.

FnrS RNA is a family of Hfq-binding small RNA whose expression is upregulated in response to anaerobic conditions. It is named FnrS because its expression is strongly dependent on fumarate and nitrate reductase regulator (FNR), a direct oxygen availability sensor.

An RNA thermometer is a temperature-sensitive non-coding RNA molecule which regulates gene expression. RNA thermometers often regulate genes required during either a heat shock or cold shock response, but have been implicated in other regulatory roles such as in pathogenicity and starvation.

In molecular biology, the ars operon is an operon found in several bacterial taxon. It is required for the detoxification of arsenate, arsenite, and antimonite. This system transports arsenite and antimonite out of the cell. The pump is composed of two polypeptides, the products of the arsA and arsB genes. This two-subunit enzyme produces resistance to arsenite and antimonite. Arsenate, however, must first be reduced to arsenite before it is extruded. A third gene, arsC, expands the substrate specificity to allow for arsenate pumping and resistance. ArsC is an approximately 150-residue arsenate reductase that uses reduced glutathione (GSH) to convert arsenate to arsenite with a redox active cysteine residue in the active site. ArsC forms an active quaternary complex with GSH, arsenate, and glutaredoxin 1 (Grx1). The three ligands must be present simultaneously for reduction to occur.
In molecular biology, the SR1 RNA is a small RNA (sRNA) produced by species of Bacillus and closely related bacteria. It is a dual-function RNA which acts both as a protein-coding RNA and as a regulatory sRNA.
Bacterial small RNAs (sRNA) are an important class of regulatory molecules in bacteria such as Brucella. They are often bound to the chaperone protein Hfq, which allows them to interact with mRNA(s). In Brucella suis 1330 RNA sequencing identified a novel list of 33 sRNAs and 62 Hfq-associated mRNAs. In Brucella melitensis eight novel sRNA genes were identified using bioinformatic and experimental approach. One of them BSR0602 was found to modulate the intracellular survival of B. melitensis. In another large-scale deep sequencing study 1321 sRNAs were identified in B. melitensis. BSR0441 sRNA was further investigated in this study and shown to play role in the intracellular survival. sRNA BM-sr0117 from Brucella melitensis was identified and shown to be bound to and cleaved by Bm-RNase III. AbcR and AbcR2 were studied B. abortus. Seven novel sRNAs were validated and their interaction with a putative target sequence was verified in B. abortus.
The tyrT operon is a series of genes encoding the tRNA for tyrosine in Escherichia coli. It is activated in response to amino acid starvation.
Colanic acid is an exopolysaccharide synthesized by bacteria in the Enterobacteriaceae family. It is excreted by the cell to form a protective bacterial capsule, and it assists in the formation of biofilms.














