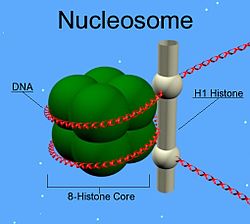
In molecular biology, linker DNA is double-stranded DNA (38-53 base pairs long) in between two nucleosome cores that, in association with histone H1, holds the cores together. Linker DNA is seen as the string in the "beads and string model", which is made by using an ionic solution on the chromatin. Linker DNA connects to histone H1 and histone H1 sits on the nucleosome core. Nucleosome is technically the consolidation of a nucleosome core and one adjacent linker DNA; however, the term nucleosome is used freely for solely the core. Linker DNA may be degraded by endonucleases. [1]
The linkers are short double stranded DNA segments which are formed of oligonucleotides. These contain target sites for the action of one or more restriction enzymes. The linkers can be synthesized chemically and can be ligated to the blunt end of foreign DNA or vector DNA. These are then treated with restriction endonuclease enzyme to produce cohesive ends of DNA fragments. The commonly used linkers are EcoRI-linkers and sal-I linkers.