In human genetics, the Y-chromosomal most recent common ancestor is the patrilineal most recent common ancestor (MRCA) from whom all currently living humans are descended. He is the most recent male from whom all living humans are descended through an unbroken line of their male ancestors. The term Y-MRCA reflects the fact that the Y chromosomes of all currently living human males are directly derived from the Y chromosome of this remote ancestor. The analogous concept of the matrilineal most recent common ancestor is known as "Mitochondrial Eve", the most recent woman from whom all living humans are descended matrilineally. As with "Mitochondrial Eve", the title of "Y-chromosomal Adam" is not permanently fixed to a single individual, but can advance over the course of human history as paternal lineages become extinct.

In genetics, a single-nucleotide polymorphism is a germline substitution of a single nucleotide at a specific position in the genome. Although certain definitions require the substitution to be present in a sufficiently large fraction of the population, many publications do not apply such a frequency threshold.

A haplotype is a group of alleles in an organism that are inherited together from a single parent.
This is a list of topics in molecular biology. See also index of biochemistry articles.
A genetic marker is a gene or DNA sequence with a known location on a chromosome that can be used to identify individuals or species. It can be described as a variation that can be observed. A genetic marker may be a short DNA sequence, such as a sequence surrounding a single base-pair change, or a long one, like minisatellites.
Indel is a molecular biology term for an insertion or deletion of bases in the genome of an organism. It is classified among small genetic variations, measuring from 1 to 10 000 base pairs in length, including insertion and deletion events that may be separated by many years, and may not be related to each other in any way. A microindel is defined as an indel that results in a net change of 1 to 50 nucleotides.

AFLP-PCR or just AFLP is a PCR-based tool used in genetics research, DNA fingerprinting, and in the practice of genetic engineering. Developed in the early 1990s by Keygene, AFLP uses restriction enzymes to digest genomic DNA, followed by ligation of adaptors to the sticky ends of the restriction fragments. A subset of the restriction fragments is then selected to be amplified. This selection is achieved by using primers complementary to the adaptor sequence, the restriction site sequence and a few nucleotides inside the restriction site fragments. The amplified fragments are separated and visualized on denaturing on agarose gel electrophoresis, either through autoradiography or fluorescence methodologies, or via automated capillary sequencing instruments.
A haplotype is a group of alleles in an organism that are inherited together from a single parent, and a haplogroup is a group of similar haplotypes that share a common ancestor with a single-nucleotide polymorphism mutation. More specifically, a haplogroup is a combination of alleles at different chromosomal regions that are closely linked and that tend to be inherited together. As a haplogroup consists of similar haplotypes, it is usually possible to predict a haplogroup from haplotypes. Haplogroups pertain to a single line of descent. As such, membership of a haplogroup, by any individual, relies on a relatively small proportion of the genetic material possessed by that individual.

In population genetics, an ancestry-informative marker (AIM) is a single-nucleotide polymorphism that exhibits substantially different frequencies between different populations. A set of many AIMs can be used to estimate the proportion of ancestry of an individual derived from each population.
A molecular marker is a molecule contained within a sample taken from an organism or other matter. It can be used to reveal certain characteristics about the respective source. DNA, for example, is a molecular marker containing information about genetic disorders and the evolutionary history of life. Specific regions of the DNA are used for diagnosing the autosomal recessive genetic disorder cystic fibrosis, taxonomic affinity (phylogenetics) and identity. Further, life forms are known to shed unique chemicals, including DNA, into the environment as evidence of their presence in a particular location. Other biological markers, like proteins, are used in diagnostic tests for complex neurodegenerative disorders, such as Alzheimer's disease. Non-biological molecular markers are also used, for example, in environmental studies.
In genetics, a subclade is a subgroup of a haplogroup.
In genetic genealogy, a unique-event polymorphism (UEP) is a genetic marker that corresponds to a mutation that is likely to occur so infrequently that it is believed overwhelmingly probable that all the individuals who share the marker, worldwide, will have inherited it from the same common ancestor, and the same single mutation event.
Haplogroup DE is a human Y-chromosome DNA haplogroup. It is defined by the single nucleotide polymorphism (SNP) mutations, or UEPs, M1(YAP), M145(P205), M203, P144, P153, P165, P167, P183. DE is unique because it is distributed in several geographically distinct clusters. An immediate subclade, haplogroup D, is mainly found in East Asia, parts of Central Asia, and the Andaman Islands, but also sporadically in West Africa and West Asia. The other immediate subclade, haplogroup E, is common in Africa, and to a lesser extent the Middle East and Europe.
Marker assisted selection or marker aided selection (MAS) is an indirect selection process where a trait of interest is selected based on a marker linked to a trait of interest, rather than on the trait itself. This process has been extensively researched and proposed for plant and animal breeding.

DNAPrint Genomics was a genetics company with a wide range of products related to genetic profiling. They were the first company to introduce forensic and consumer genomics products, which were developed immediately upon the publication of the first complete draft of the human genome in the early 2000s. They researched, developed, and marketed the first ever consumer genomics product, based on "Ancestry Informative Markers" which they used to correctly identify the BioGeographical Ancestry (BGA) of a human based on a sample of their DNA. They also researched, developed and marketed the first ever forensic genomics product - DNAWITNESS - which was used to create a physical profile of donors of crime scene DNA. The company reached a peak of roughly $3M/year revenues but ceased operations in February 2009.
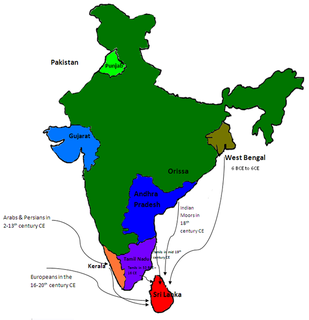
Genetic studies on the Sinhalese is part of population genetics investigating the origins of the Sinhalese population.
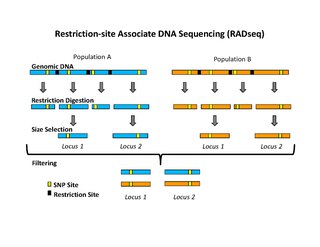
Restriction site associated DNA (RAD) markers are a type of genetic marker which are useful for association mapping, QTL-mapping, population genetics, ecological genetics and evolutionary genetics. The use of RAD markers for genetic mapping is often called RAD mapping. An important aspect of RAD markers and mapping is the process of isolating RAD tags, which are the DNA sequences that immediately flank each instance of a particular restriction site of a restriction enzyme throughout the genome. Once RAD tags have been isolated, they can be used to identify and genotype DNA sequence polymorphisms mainly in form of single nucleotide polymorphisms (SNPs). Polymorphisms that are identified and genotyped by isolating and analyzing RAD tags are referred to as RAD markers.

A gene is said to be polymorphic if more than one allele occupies that gene's locus within a population. In addition to having more than one allele at a specific locus, each allele must also occur in the population at a rate of at least 1% to generally be considered polymorphic.
Although Sri Lankan Tamils are culturally and linguistically distinct, genetic studies indicate that they are closely related to other ethnic groups in the island while being related to the Indian Tamils from South India and Bengalis from the East India as well. There are various studies that indicate varying degrees of connections between Sri Lankan Tamils, Sinhalese and Indian ethnic groups.