Related Research Articles
Functional integration is the study of how brain regions work together to process information and effect responses. Though functional integration frequently relies on anatomic knowledge of the connections between brain areas, the emphasis is on how large clusters of neurons – numbering in the thousands or millions – fire together under various stimuli. The large datasets required for such a whole-scale picture of brain function have motivated the development of several novel and general methods for the statistical analysis of interdependence, such as dynamic causal modelling and statistical linear parametric mapping. These datasets are typically gathered in human subjects by non-invasive methods such as EEG/MEG, fMRI, or PET. The results can be of clinical value by helping to identify the regions responsible for psychiatric disorders, as well as to assess how different activities or lifestyles affect the functioning of the brain.

Functional near-infrared spectroscopy (fNIRS) is an optical brain monitoring technique which uses near-infrared spectroscopy for the purpose of functional neuroimaging. Using fNIRS, brain activity is measured by using near-infrared light to estimate cortical hemodynamic activity which occur in response to neural activity. Alongside EEG, fNIRS is one of the most common non-invasive neuroimaging techniques which can be used in portable contexts. The signal is often compared with the BOLD signal measured by fMRI and is capable of measuring changes both in oxy- and deoxyhemoglobin concentration, but can only measure from regions near the cortical surface. fNIRS may also be referred to as Optical Topography (OT) and is sometimes referred to simply as NIRS.
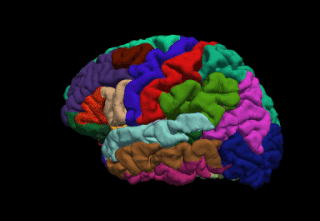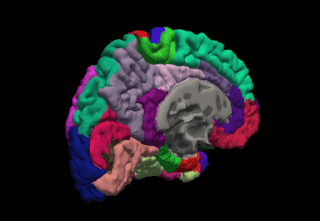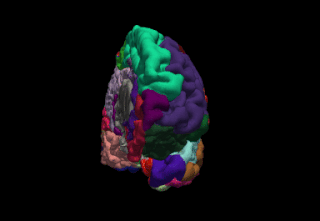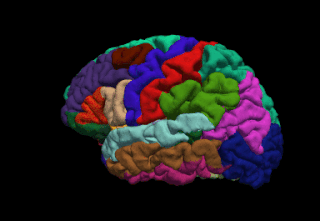
FreeSurfer is a brain imaging software package originally developed by Bruce Fischl, Anders Dale, Martin Sereno, and Doug Greve. Development and maintenance of FreeSurfer is now the primary responsibility of the Laboratory for Computational Neuroimaging at the Athinoula A. Martinos Center for Biomedical Imaging. FreeSurfer contains a set of programs with a common focus of analyzing magnetic resonance imaging (MRI) scans of brain tissue. It is an important tool in functional brain mapping and contains tools to conduct both volume based and surface based analysis. FreeSurfer includes tools for the reconstruction of topologically correct and geometrically accurate models of both the gray/white and pial surfaces, for measuring cortical thickness, surface area and folding, and for computing inter-subject registration based on the pattern of cortical folds.
Connectomics is the production and study of connectomes: comprehensive maps of connections within an organism's nervous system. More generally, it can be thought of as the study of neuronal wiring diagrams with a focus on how structural connectivity, individual synapses, cellular morphology, and cellular ultrastructure contribute to the make up of a network. The nervous system is a network made of billions of connections and these connections are responsible for our thoughts, emotions, actions, memories, function and dysfunction. Therefore, the study of connectomics aims to advance our understanding of mental health and cognition by understanding how cells in the nervous system are connected and communicate. Because these structures are extremely complex, methods within this field use a high-throughput application of functional and structural neural imaging, most commonly magnetic resonance imaging (MRI), electron microscopy, and histological techniques in order to increase the speed, efficiency, and resolution of these nervous system maps. To date, tens of large scale datasets have been collected spanning the nervous system including the various areas of cortex, cerebellum, the retina, the peripheral nervous system and neuromuscular junctions.

In neuroscience, the default mode network (DMN), also known as the default network, default state network, or anatomically the medial frontoparietal network (M-FPN), is a large-scale brain network primarily composed of the dorsal medial prefrontal cortex, posterior cingulate cortex, precuneus and angular gyrus. It is best known for being active when a person is not focused on the outside world and the brain is at wakeful rest, such as during daydreaming and mind-wandering. It can also be active during detailed thoughts related to external task performance. Other times that the DMN is active include when the individual is thinking about others, thinking about themselves, remembering the past, and planning for the future.
Psychophysiological interaction (PPI) is a brain connectivity analysis method for functional brain imaging data, mainly functional magnetic resonance imaging (fMRI). It estimates context-dependent changes in effective connectivity (coupling) between brain regions. Thus, PPI analysis identifies brain regions whose activity depends on an interaction between psychological context and physiological state of the seed region.
The Human Connectome Project (HCP) is a five-year project sponsored by sixteen components of the National Institutes of Health, split between two consortia of research institutions. The project was launched in July 2009 as the first of three Grand Challenges of the NIH's Blueprint for Neuroscience Research. On September 15, 2010, the NIH announced that it would award two grants: $30 million over five years to a consortium led by Washington University in St. Louis and the University of Minnesota, with strong contributions from University of Oxford (FMRIB) and $8.5 million over three years to a consortium led by Harvard University, Massachusetts General Hospital and the University of California Los Angeles.
Anders Martin Dale is a prominent neuroscientist and professor of radiology, neurosciences, psychiatry, and cognitive science at the University of California, San Diego (UCSD), and is one of the world's leading developers of sophisticated computational neuroimaging techniques. He is the founding Director of the Center for Multimodal Imaging Genetics (CMIG) at UCSD.

Resting state fMRI is a method of functional magnetic resonance imaging (fMRI) that is used in brain mapping to evaluate regional interactions that occur in a resting or task-negative state, when an explicit task is not being performed. A number of resting-state brain networks have been identified, one of which is the default mode network. These brain networks are observed through changes in blood flow in the brain which creates what is referred to as a blood-oxygen-level dependent (BOLD) signal that can be measured using fMRI.
Dynamic functional connectivity (DFC) refers to the observed phenomenon that functional connectivity changes over a short time. Dynamic functional connectivity is a recent expansion on traditional functional connectivity analysis which typically assumes that functional networks are static in time. DFC is related to a variety of different neurological disorders, and has been suggested to be a more accurate representation of functional brain networks. The primary tool for analyzing DFC is fMRI, but DFC has also been observed with several other mediums. DFC is a recent development within the field of functional neuroimaging whose discovery was motivated by the observation of temporal variability in the rising field of steady state connectivity research.

CONN is a Matlab-based cross-platform imaging software for the computation, display, and analysis of functional connectivity in fMRI in the resting state and during task.
Dynamic causal modeling (DCM) is a framework for specifying models, fitting them to data and comparing their evidence using Bayesian model comparison. It uses nonlinear state-space models in continuous time, specified using stochastic or ordinary differential equations. DCM was initially developed for testing hypotheses about neural dynamics. In this setting, differential equations describe the interaction of neural populations, which directly or indirectly give rise to functional neuroimaging data e.g., functional magnetic resonance imaging (fMRI), magnetoencephalography (MEG) or electroencephalography (EEG). Parameters in these models quantify the directed influences or effective connectivity among neuronal populations, which are estimated from the data using Bayesian statistical methods.

John-Dylan Haynes is a British-German brain researcher.
Brain–body interactions are patterns of neural activity in the central nervous system to coordinate the activity between the brain and body. The nervous system consists of central and peripheral nervous systems and coordinates the actions of an animal by transmitting signals to and from different parts of its body. The brain and spinal cord are interwoven with the body and interact with other organ systems through the somatic, autonomic and enteric nervous systems. Neural pathways regulate brain–body interactions and allow to sense and control its body and interact with the environment.
Arterial spin labeling (ASL), also known as arterial spin tagging, is a magnetic resonance imaging technique used to quantify cerebral blood perfusion by labelling blood water as it flows throughout the brain. ASL specifically refers to magnetic labeling of arterial blood below or in the imaging slab, without the need of gadolinium contrast. A number of ASL schemes are possible, the simplest being flow alternating inversion recovery (FAIR) which requires two acquisitions of identical parameters with the exception of the out-of-slice saturation; the difference in the two images is theoretically only from inflowing spins, and may be considered a 'perfusion map'. The ASL technique was developed by Alan P. Koretsky, Donald S. Williams, John A. Detre and John S. Leigh Jr in 1992.

Dimitri Van De Ville is a Swiss and Belgian computer scientist and neuroscientist specialized in dynamical and network aspects of brain activity. He is a professor of bioengineering at EPFL and the head of the Medical Image Processing Laboratory at EPFL's School of Engineering.

Alfonso Nieto-Castanon is a Spanish computational neuroscientist and developer of computational neuroimaging analysis methods and tools. He is a visiting researcher at the Boston University College of Health and Rehabilitation Sciences, and research affiliate at MIT McGovern Institute for Brain Research. His research focuses on the understanding and characterization of human brain dynamics underlying mental function.
Susan Whitfield-Gabrieli is an American scientist, psychologist/neuroscientist, academic and researcher. She is a professor of psychology, the Founding Director of the Biomedical Imaging Center at Northeastern University, Researcher in the Department of Psychiatry at Massachusetts General Hospital, Harvard Medical School and a Research Affiliate of McGovern Institute for Brain Research at Massachusetts Institute of Technology.
References
- ↑ Whitfield-Gabrieli, S.; Nieto-Castanon, A. (2012). "Conn: a functional connectivity toolbox for correlated and anticorrelated brainnetworks". Brain Connect. 2 (3): 125–141. doi:10.1089/brain.2012.0073. PMID 22642651.
- ↑ Friston, K. J.; Kahan, J.; Biswal, B.; Razi, A. (2014). "A DCM for resting state fMRI". NeuroImage. 94 (100): 396–407. doi:10.1016/j.neuroimage.2013.12.009. PMC 4073651 . PMID 24345387.
- ↑ Taylor, P. A.; Saad, Z. S. (2013). "FATCAT: (an efficient) Functional and Tractographic Connectivity Analysis Toolbox". Brain Connect. 3 (5): 523–535. doi:10.1089/brain.2013.0154. PMC 3796333 . PMID 23980912.
- ↑ Fischl, B. FreeSurfer (2012). "FreeSurfer". NeuroImage. 62 (2): 774–781. doi:10.1016/j.neuroimage.2012.01.021. PMC 3685476 . PMID 22248573.
- ↑ Beckmann, C. F.; DeLuca, M.; Devlin, J. T.; Smith, S. M. (2005). "Investigations into resting-state connectivity using independentcomponent analysis". Philos. Trans. R. Soc. Lond. B Biol. Sci. 360 (1457): 1001–1013. doi:10.1098/rstb.2005.1634. PMC 1854918 . PMID 16087444.