
Escherichia coli, also known as E. coli, is a Gram-negative, facultative anaerobic, rod-shaped, coliform bacterium of the genus Escherichia that is commonly found in the lower intestine of warm-blooded organisms. Most E. coli strains are harmless, but some serotypes (EPEC, ETEC etc.) can cause serious food poisoning in their hosts, and are occasionally responsible for food contamination incidents that prompt product recalls. The harmless strains are part of the normal microbiota of the gut, and can benefit their hosts by producing vitamin K2, and preventing colonisation of the intestine with pathogenic bacteria, having a mutualistic relationship. E. coli is expelled into the environment within fecal matter. The bacterium grows massively in fresh fecal matter under aerobic conditions for three days, but its numbers decline slowly afterwards.

The human microbiome is the aggregate of all microbiota that reside on or within human tissues and biofluids along with the corresponding anatomical sites in which they reside, including the skin, mammary glands, seminal fluid, uterus, ovarian follicles, lung, saliva, oral mucosa, conjunctiva, biliary tract, and gastrointestinal tract. Types of human microbiota include bacteria, archaea, fungi, protists and viruses. Though micro-animals can also live on the human body, they are typically excluded from this definition. In the context of genomics, the term human microbiome is sometimes used to refer to the collective genomes of resident microorganisms; however, the term human metagenome has the same meaning.
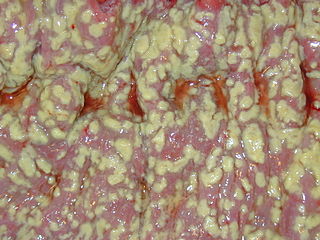
Clostridioides difficile infection , also known as Clostridium difficile infection, is a symptomatic infection due to the spore-forming bacterium Clostridioides difficile. Symptoms include watery diarrhea, fever, nausea, and abdominal pain. It makes up about 20% of cases of antibiotic-associated diarrhea. Antibiotics can contribute to detrimental changes in gut microbiota; specifically, they decrease short-chain fatty acid absorption which results in osmotic, or watery, diarrhea. Complications may include pseudomembranous colitis, toxic megacolon, perforation of the colon, and sepsis.

The Clostridia are a highly polyphyletic class of Bacillota, including Clostridium and other similar genera. They are distinguished from the Bacilli by lacking aerobic respiration. They are obligate anaerobes and oxygen is toxic to them. Species of the class Clostridia are often but not always Gram-positive and have the ability to form spores. Studies show they are not a monophyletic group, and their relationships are not entirely certain. Currently, most are placed in a single order called Clostridiales, but this is not a natural group and is likely to be redefined in the future.

Coliform bacteria are defined as either motile or non-motile Gram-negative non-spore forming Bacilli that possess β-galactosidase to produce acids and gases under their optimal growth temperature of 35-37°C. They can be aerobes or facultative aerobes, and are a commonly used indicator of low sanitary quality of foods, milk, and water. Coliforms can be found in the aquatic environment, in soil and on vegetation; they are universally present in large numbers in the feces of warm-blooded animals as they are known to inhabit the gastrointestinal system. While coliform bacteria are not normally causes of serious illness, they are easy to culture, and their presence is used to infer that other pathogenic organisms of fecal origin may be present in a sample, or that said sample is not safe to consume. Such pathogens include disease-causing bacteria, viruses, or protozoa and many multicellular parasites.

An opportunistic infection is an infection caused by pathogens that take advantage of an opportunity not normally available. These opportunities can stem from a variety of sources, such as a weakened immune system, an altered microbiome, or breached integumentary barriers. Many of these pathogens do not necessarily

Gutmicrobiota are the microorganisms, including bacteria and archaea, that live in the digestive tracts of vertebrates including humans, and of insects. Alternative terms include gutflora and gutmicrobiome. The gastrointestinal metagenome is the aggregate of all the genomes of gut microbiota. In the human, the gut is the main location of human microbiota. The gut microbiota has broad impacts, including effects on colonization, resistance to pathogens, maintaining the intestinal epithelium, metabolizing dietary and pharmaceutical compounds, controlling immune function, and even behavior through the gut-brain axis.

Fecal microbiota transplant (FMT), also known as a stool transplant, is the process of transferring fecal bacteria and other microbes from a healthy individual into another individual. FMT is an effective treatment for Clostridioides difficile infection (CDI). For recurrent CDI, FMT is more effective than vancomycin.
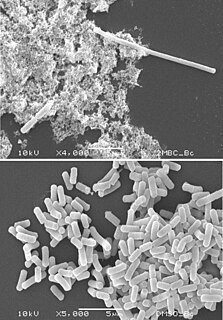
Filamentation, also termed conditional filamentation, is the anomalous growth of certain bacteria, such as Escherichia coli, in which cells continue to elongate but do not divide. The cells that result from elongation without division have multiple chromosomal copies. In the absence of antibiotics or other stressors, filamentation occurs at a low frequency in bacterial populations. The increased cell length can protect bacteria from protozoan predation and neutrophil phagocytosis by making ingestion of cells more difficult. Filamentation is also thought to protect bacteria from antibiotics, and is associated with other aspects of bacterial virulence such as biofilm formation. The number and length of filaments within a bacterial population increases when the bacteria are treated with various chemical and physical agents. Some of the key genes involved in filamentation in E. coli include sulA and minCD.
Dysbiosis is characterized as a disruption to the microbiota homeostasis caused by an imbalance in the microflora, changes in their functional composition and metabolic activities, or a shift in their local distribution. It is a term for a microbial imbalance or maladaptation on or inside the body, such as an impaired microbiota. For example, a part of the human microbiota, such as the skin flora, gut flora, or vaginal flora, can become deranged, with normally dominating species underrepresented and normally outcompeted or contained species increasing to fill the void. Dysbiosis is most commonly reported as a condition in the gastrointestinal tract, particularly during small intestinal bacterial overgrowth (SIBO) or small intestinal fungal overgrowth (SIFO).
Saccharomyces boulardii is a tropical yeast first isolated from lychee and mangosteen fruit peel in 1923 by French scientist Henri Boulard. Although early reports claimed distinct taxonomic, metabolic, and genetic properties, S. boulardii is genetically a grouping of S. cerevisiae strains, sharing >99% genomic relatedness, giving the synonym S. cerevisiae var. boulardii.

Human feces is the solid or semisolid remains of food that could not be digested or absorbed in the small intestine of humans, but has been further broken down by bacteria in the large intestine. It also contains bacteria and a relatively small amount of metabolic waste products such as bacterially altered bilirubin, and the dead epithelial cells from the lining of the gut. It is discharged through the anus during a process called defecation.
Clostridium innocuum is an anaerobic, non-motile, gram-positive bacterium that reproduces by sporulation. While there are over 130 species of Clostridium, C. innocuum is the third most commonly isolated. Although it is not normally considered an aggressive human pathogen, it has been isolated in some disease processes. C. innocuum and other Clostridium line the oropharynx and gastrointestinal tract, and are considered normal gut flora.
Clostridium cadaveris is an enteric, gas-forming, motile, strictly anaerobic gram-positive bacterium of the genus Clostridium. First described by Klein in 1899, it was noted to be the most prominent bacteria during human decomposition; historically it was described as "putrefying flora".

Escherichia coli is a gram-negative, rod-shaped bacterium that is commonly found in the lower intestine of warm-blooded organisms (endotherms). Most E. coli strains are harmless, but pathogenic varieties cause serious food poisoning, septic shock, meningitis, or urinary tract infections in humans. Unlike normal flora E. coli, the pathogenic varieties produce toxins and other virulence factors that enable them to reside in parts of the body normally not inhabited by E. coli, and to damage host cells. These pathogenic traits are encoded by virulence genes carried only by the pathogens.
Clostridium leptum is a bacterium species in the genus Clostridium.
Bacteriotherapy is the purposeful use of bacteria or their products in treating an illness. Forms of bacteriotherapy include the use of probiotics, microorganisms that provide health benefits when consumed; fecal matter transplants (FMT) /intestinal microbiota transplant (IMT), the transfer of gut microorganisms from the fecal matter of healthy donors to recipient patients to restore microbiota; or synbiotics which combine prebiotics, indigestible ingredients that promote growth of beneficial microorganisms, and probiotics. Through these methods, the gut microbiota, the community of 300-500 microorganism species that live in the digestive tract of animals aiding in digestion, energy storage, immune function and protection against pathogens, can be recolonized with favorable bacteria, which in turn has therapeutic effects.

Clostridioides difficile, also known as C. difficile, or C. diff, is Gram-positive species of spore-forming bacteria. Clostridioides spp. are anaerobic, motile bacteria, ubiquitous in nature and especially prevalent in soil. Its vegetative cells are rod-shaped, pleomorphic, and occur in pairs or short chains. Under the microscope, they appear as long, irregular cells with a bulge at their terminal ends. Under Gram staining, C. difficile cells are Gram-positive and show optimum growth on blood agar at human body temperatures in the absence of oxygen. C. difficile is catalase- and superoxide dismutase-negative, and produces up to three types of toxins: enterotoxin A, cytotoxin B and Cytolethal distending toxin. Under stress conditions, the bacteria produce spores that are able to tolerate extreme conditions that the active bacteria cannot tolerate.
Proteobiotics are natural metabolites which are produced by fermentation process of specific probiotic strains. These small oligopeptides were originally discovered in and isolated from culture media used to grow probiotic bacteria and may account for some of the health benefits of probiotics.
Dactylosporangium aurantiacum is a Gram-positive soil-based actinobacterium in the family Micromonosporaceae. Like all Dactylosporangium species, aurantiacum is aerobic and mesophilic.









