
An antibiotic is a type of antimicrobial substance active against bacteria. It is the most important type of antibacterial agent for fighting bacterial infections, and antibiotic medications are widely used in the treatment and prevention of such infections. They may either kill or inhibit the growth of bacteria. A limited number of antibiotics also possess antiprotozoal activity. Antibiotics are not effective against viruses such as the ones which cause the common cold or influenza; drugs which inhibit growth of viruses are termed antiviral drugs or antivirals rather than antibiotics. They are also not effective against fungi; drugs which inhibit growth of fungi are called antifungal drugs.

Beta-lactamases (β-lactamases) are enzymes produced by bacteria that provide multi-resistance to beta-lactam antibiotics such as penicillins, cephalosporins, cephamycins, monobactams and carbapenems (ertapenem), although carbapenems are relatively resistant to beta-lactamase. Beta-lactamase provides antibiotic resistance by breaking the antibiotics' structure. These antibiotics all have a common element in their molecular structure: a four-atom ring known as a beta-lactam (β-lactam) ring. Through hydrolysis, the enzyme lactamase breaks the β-lactam ring open, deactivating the molecule's antibacterial properties.

Escherichia coli ( ESH-ə-RIK-ee-ə KOH-lye) is a gram-negative, facultative anaerobic, rod-shaped, coliform bacterium of the genus Escherichia that is commonly found in the lower intestine of warm-blooded organisms. Most E. coli strains are harmless, but some serotypes such as EPEC, and ETEC are pathogenic and can cause serious food poisoning in their hosts, and are occasionally responsible for food contamination incidents that prompt product recalls. Most strains are part of the normal microbiota of the gut and are harmless or even beneficial to humans (although these strains tend to be less studied than the pathogenic ones). For example, some strains of E. coli benefit their hosts by producing vitamin K2 or by preventing the colonization of the intestine by pathogenic bacteria. These mutually beneficial relationships between E. coli and humans are a type of mutualistic biological relationship — where both the humans and the E. coli are benefitting each other. E. coli is expelled into the environment within fecal matter. The bacterium grows massively in fresh fecal matter under aerobic conditions for three days, but its numbers decline slowly afterwards.

A flagellum is a hairlike appendage that protrudes from certain plant and animal sperm cells, from fungal spores (zoospores), and from a wide range of microorganisms to provide motility. Many protists with flagella are known as flagellates.

Horizontal gene transfer (HGT) or lateral gene transfer (LGT) is the movement of genetic material between organisms other than by the ("vertical") transmission of DNA from parent to offspring (reproduction). HGT is an important factor in the evolution of many organisms. HGT is influencing scientific understanding of higher-order evolution while more significantly shifting perspectives on bacterial evolution.

Geobiology is a field of scientific research that explores the interactions between the physical Earth and the biosphere. It is a relatively young field, and its borders are fluid. There is considerable overlap with the fields of ecology, evolutionary biology, microbiology, paleontology, and particularly soil science and biogeochemistry. Geobiology applies the principles and methods of biology, geology, and soil science to the study of the ancient history of the co-evolution of life and Earth as well as the role of life in the modern world. Geobiologic studies tend to be focused on microorganisms, and on the role that life plays in altering the chemical and physical environment of the pedosphere, which exists at the intersection of the lithosphere, atmosphere, hydrosphere and/or cryosphere. It differs from biogeochemistry in that the focus is on processes and organisms over space and time rather than on global chemical cycles.

Carl Zimmer is a popular science writer, blogger, columnist, and journalist who specializes in the topics of evolution, parasites, and heredity. The author of many books, he contributes science essays to publications such as The New York Times, Discover, and National Geographic. He is a fellow at Yale University's Morse College and adjunct professor of molecular biophysics and biochemistry at Yale University. Zimmer also gives frequent lectures and has appeared on many radio shows, including National Public Radio's Radiolab, Fresh Air, and This American Life.

Richard Eimer Lenski is an American evolutionary biologist, a Hannah Distinguished Professor of Microbial Ecology, Genetics and Evolution, and Evolution of Pathogen Virulence at Michigan State University. He is a member of the National Academy of Sciences and a MacArthur Fellow. Lenski is best known for his still ongoing 36-year-old long-term E. coli evolution experiment, which has been instrumental in understanding the core processes of evolution, including mutation rates, clonal interference, antibiotic resistance, the evolution of novel traits, and speciation. He is also well known for his pioneering work in studying evolution digitally using self-replicating organisms called Avida.
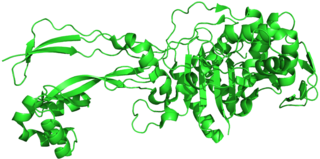
Penicillin-binding proteins (PBPs) are a group of proteins that are characterized by their affinity for and binding of penicillin. They are a normal constituent of many bacteria; the name just reflects the way by which the protein was discovered. All β-lactam antibiotics bind to PBPs, which are essential for bacterial cell wall synthesis. PBPs are members of a subgroup of enzymes called transpeptidases. Specifically, PBPs are DD-transpeptidases.

A chemostat is a bioreactor to which fresh medium is continuously added, while culture liquid containing left over nutrients, metabolic end products and microorganisms is continuously removed at the same rate to keep the culture volume constant. By changing the rate with which medium is added to the bioreactor the specific growth rate of the microorganism can be easily controlled within limits.
A shadow biosphere is a hypothetical microbial biosphere of Earth that would use radically different biochemical and molecular processes from that of currently known life. Although life on Earth is relatively well studied, if a shadow biosphere exists, it may still remain unnoticed because the exploration of the microbial world targets primarily the biochemistry of the macro-organisms.
Allan McCulloch Campbell was an American microbiologist and geneticist and the Barbara Kimball Browning Professor Emeritus in the Department of Biology at Stanford University. His pioneering work on Lambda phage helped to advance molecular biology in the late 20th century. An important collaborator and member of his laboratory at Stanford University was biochemist Alice del Campillo Campbell, his wife.

Esther Miriam Zimmer Lederberg was an American microbiologist and a pioneer of bacterial genetics. She discovered the bacterial virus lambda phage and the bacterial fertility factor F, devised the first implementation of replica plating, and furthered the understanding of the transfer of genes between bacteria by specialized transduction.

Bacteria are ubiquitous, mostly free-living organisms often consisting of one biological cell. They constitute a large domain of prokaryotic microorganisms. Typically a few micrometres in length, bacteria were among the first life forms to appear on Earth, and are present in most of its habitats. Bacteria inhabit soil, water, acidic hot springs, radioactive waste, and the deep biosphere of Earth's crust. Bacteria play a vital role in many stages of the nutrient cycle by recycling nutrients and the fixation of nitrogen from the atmosphere. The nutrient cycle includes the decomposition of dead bodies; bacteria are responsible for the putrefaction stage in this process. In the biological communities surrounding hydrothermal vents and cold seeps, extremophile bacteria provide the nutrients needed to sustain life by converting dissolved compounds, such as hydrogen sulphide and methane, to energy. Bacteria also live in mutualistic, commensal and parasitic relationships with plants and animals. Most bacteria have not been characterised and there are many species that cannot be grown in the laboratory. The study of bacteria is known as bacteriology, a branch of microbiology.

Parasite Rex: Inside the Bizarre World of Nature's Most Dangerous Creatures is a nonfiction book by Carl Zimmer that was published by Free Press in 2000. The book discusses the history of parasites on Earth and how the field and study of parasitology formed, along with a look at the most dangerous parasites ever found in nature. A special paperback edition was released in March 2011 for the tenth anniversary of the book's publishing, including a new epilogue written by Zimmer. Signed bookplates were also given to fans that sent in a photo of themselves with a copy of the special edition.
Mycoplasma laboratorium or Synthia refers to a synthetic strain of bacterium. The project to build the new bacterium has evolved since its inception. Initially the goal was to identify a minimal set of genes that are required to sustain life from the genome of Mycoplasma genitalium, and rebuild these genes synthetically to create a "new" organism. Mycoplasma genitalium was originally chosen as the basis for this project because at the time it had the smallest number of genes of all organisms analyzed. Later, the focus switched to Mycoplasma mycoides and took a more trial-and-error approach.

Microbiology is the scientific study of microorganisms, those being of unicellular (single-celled), multicellular, or acellular. Microbiology encompasses numerous sub-disciplines including virology, bacteriology, protistology, mycology, immunology, and parasitology.
Pathogenomics is a field which uses high-throughput screening technology and bioinformatics to study encoded microbe resistance, as well as virulence factors (VFs), which enable a microorganism to infect a host and possibly cause disease. This includes studying genomes of pathogens which cannot be cultured outside of a host. In the past, researchers and medical professionals found it difficult to study and understand pathogenic traits of infectious organisms. With newer technology, pathogen genomes can be identified and sequenced in a much shorter time and at a lower cost, thus improving the ability to diagnose, treat, and even predict and prevent pathogenic infections and disease. It has also allowed researchers to better understand genome evolution events - gene loss, gain, duplication, rearrangement - and how those events impact pathogen resistance and ability to cause disease. This influx of information has created a need for bioinformatics tools and databases to analyze and make the vast amounts of data accessible to researchers, and it has raised ethical questions about the wisdom of reconstructing previously extinct and deadly pathogens in order to better understand virulence.

Escherichia coli is a gram-negative, rod-shaped bacterium that is commonly found in the lower intestine of warm-blooded organisms (endotherms). Most E. coli strains are harmless, but pathogenic varieties cause serious food poisoning, septic shock, meningitis, or urinary tract infections in humans. Unlike normal flora E. coli, the pathogenic varieties produce toxins and other virulence factors that enable them to reside in parts of the body normally not inhabited by E. coli, and to damage host cells. These pathogenic traits are encoded by virulence genes carried only by the pathogens.
The host–pathogen interaction is defined as how microbes or viruses sustain themselves within host organisms on a molecular, cellular, organismal or population level. This term is most commonly used to refer to disease-causing microorganisms although they may not cause illness in all hosts. Because of this, the definition has been expanded to how known pathogens survive within their host, whether they cause disease or not.













