
Nanoarchaeota is a proposed phylum in the domain Archaea that currently has only one representative, Nanoarchaeum equitans, which was discovered in a submarine hydrothermal vent and first described in 2002.

Gracilicutes is a clade in bacterial phylogeny.

The PVC superphylum is a superphylum of bacteria named after its three important members, Planctomycetota, Verrucomicrobiota, and Chlamydiota. Cavalier-Smith postulated that the PVC bacteria probably lost or reduced their peptidoglycan cell wall twice. It has been hypothesised that a member of the PVC clade might have been the host cell in the endosymbiotic event that gave rise to the first proto-eukaryotic cell.

Terrabacteria is a taxon containing approximately two-thirds of prokaryote species, including those in the gram positive phyla as well as the phyla "Cyanobacteria", Chloroflexota, and Deinococcota.

The FCB group is a superphylum of bacteria named after the main member phyla Fibrobacterota, Chlorobiota, and Bacteroidota. The members are considered to form a clade due to a number of conserved signature indels.
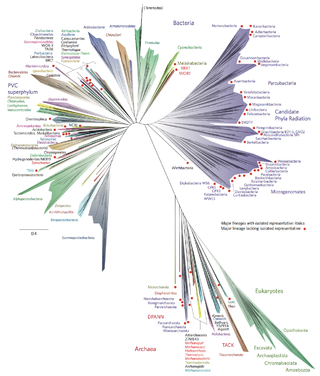
Bacterial phyla constitute the major lineages of the domain Bacteria. While the exact definition of a bacterial phylum is debated, a popular definition is that a bacterial phylum is a monophyletic lineage of bacteria whose 16S rRNA genes share a pairwise sequence identity of ~75% or less with those of the members of other bacterial phyla.
Saccharibacteria, formerly known as TM7, is a major bacterial lineage. It was discovered through 16S rRNA sequencing.

Hydrobacteria is a taxon containing approximately one-third of prokaryote species, mostly gram-negative bacteria and their relatives. It was found to be the closest relative of an even larger group of Bacteria, Terrabacteria, which are mostly gram positive bacteria. The name Hydrobacteria refers to the moist environment inferred for the common ancestor of those species. In contrast, species of Terrabacteria possess adaptations for life on land.

DPANN is a superphylum of Archaea first proposed in 2013. Many members show novel signs of horizontal gene transfer from other domains of life. They are known as nanoarchaea or ultra-small archaea due to their smaller size (nanometric) compared to other archaea.

The Orthokaryotes are a proposed Eukaryote clade consisting of the Jakobea and the Neokaryotes. Together with its sister Discicristata it forms a basal Eukaryote clade. They are characterized by stacked Golgi, orthogonal centrioles, and two opposite posterior ciliary roots.

The neokaryotes are a proposed eukaryote clade consisting of the unikonts and the bikonts as sister of for instance the Jakobea. It arises because the Euglenozoa, Percolozoa, Tsukubea, and Jakobea are seen in this view as more basal eukaryotes. These four groups, are traditionally grouped together in the Discoba. However, the Discoba may well be paraphyletic as the neokaryotes may have emerged in them.
The Scotokaryotes (Cavalier-Smith) is a proposed basal Neokaryote clade as sister of the Diaphoretickes. Basal Scotokaryote groupings are the Metamonads, the Malawimonas and the Podiata. In this phylogeny the Discoba are sometimes seen as paraphyletic and basal Eukaryotes.
The candidate phyla radiation is a large evolutionary radiation of bacterial lineages whose members are mostly uncultivated and only known from metagenomics and single cell sequencing. They have been described as nanobacteria or ultra-small bacteria due to their reduced size (nanometric) compared to other bacteria.
Gracilibacteria is a bacterial candidate phylum formerly known as GN02, BD1-5, or SN-2. It is part of the Candidate Phyla Radiation and the Patescibacteria group.
Zixibacteria is a bacterial phylum with candidate status, meaning it had no cultured representatives. It is a member of the FCB group
Katanobacteria is a bacterial phylum formerly known as WWE3. It has candidate status, meaning there are no cultured representatives, and is a member of the Candidate Phyla Radiation (CPR).
Berkelbacteria is a bacterial phylum with candidate status, meaning there are no cultured representatives for this group. It is part of the Candidate Phyla Radiation.
Fertabacteria is a candidate bacterial phylum of the Candidate Phyla Radiation, first proposed in 2017 after analysis of a genome from the mouth of a bottlenose dolphin. Members of this phylum are predicted to have been widely under-detected in 16S rRNA gene-based surveys of community composition due to mismatches between commonly used primers and the corresponding primer site. Fertabacteria have been retroactively detected in a variety of environments.
CandidatusWirthbacteria is a proposed bacterial phylum containing only one known sample from the Crystal Geyser aquifer, Ca. Wirthibacter wanneri. This bacterium stands out in a basal position in some trees of life as it is closely related to Candidate phyla radiation but is not considered part of that clade.










