Related Research Articles
Insulin resistance (IR) is a pathological condition in which cells in insulin-sensitive tissues in the body fail to respond normally to the hormone insulin or downregulate insulin receptors in response to hyperinsulinemia.

The blood sugar level, blood sugar concentration, blood glucose level, or glycemia is the measure of glucose concentrated in the blood. The body tightly regulates blood glucose levels as a part of metabolic homeostasis.

Ischemia or ischaemia is a restriction in blood supply to any tissue, muscle group, or organ of the body, causing a shortage of oxygen that is needed for cellular metabolism. Ischemia is generally caused by problems with blood vessels, with resultant damage to or dysfunction of tissue i.e. hypoxia and microvascular dysfunction. It also implies local hypoxia in a part of a body resulting from constriction.
Glucokinase is an enzyme that facilitates phosphorylation of glucose to glucose-6-phosphate. Glucokinase occurs in cells in the liver and pancreas of humans and most other vertebrates. In each of these organs it plays an important role in the regulation of carbohydrate metabolism by acting as a glucose sensor, triggering shifts in metabolism or cell function in response to rising or falling levels of glucose, such as occur after a meal or when fasting. Mutations of the gene for this enzyme can cause unusual forms of diabetes or hypoglycemia.

In molecular biology, biochips are engineered substrates that can host large numbers of simultaneous biochemical reactions. One of the goals of biochip technology is to efficiently screen large numbers of biological analytes, with potential applications ranging from disease diagnosis to detection of bioterrorism agents. For example, digital microfluidic biochips are under investigation for applications in biomedical fields. In a digital microfluidic biochip, a group of (adjacent) cells in the microfluidic array can be configured to work as storage, functional operations, as well as for transporting fluid droplets dynamically.

A pulmonary artery catheter (PAC), also known as a Swan-Ganz catheter or right heart catheter, is a balloon-tipped catheter that is inserted into a pulmonary artery in a procedure known as pulmonary artery catheterization or right heart catheterization. Pulmonary artery catheterization is a useful measure of the overall function of the heart particularly in those with complications from heart failure, heart attack, arrhythmias or pulmonary embolism. It is also a good measure for those needing intravenous fluid therapy, for instance post heart surgery, shock, and severe burns. The procedure can also be used to measure pressures in the heart chambers.
In oncology, the Warburg effect is the observation that most cancer use aerobic glycolysis for energy generation rather than the mechanisms used by non-cancerous cells. This observation was first published by Otto Heinrich Warburg, who was awarded the 1931 Nobel Prize in Physiology for his "discovery of the nature and mode of action of the respiratory enzyme". The existence of the Warburg effect has fuelled popular misconceptions that cancer can be treated by dietary reductions in sugar and carbohydrate.
Machine perfusion (MP) is an artificial perfusion technique often used for organ preservation to help facilitate organ transplantation. MP works by continuously pumping a specialized solution through donor organs, mimicking the body's natural blood flow while actively controlling temperature, oxygen levels, chemical composition, and mechanical stress within the organ. By maintaining organ viability outside the body for extended periods, machine perfusion addresses critical challenges in organ transplantation, such as limited preservation times.
Respirometry is a general term that encompasses a number of techniques for obtaining estimates of the rates of metabolism of vertebrates, invertebrates, plants, tissues, cells, or microorganisms via an indirect measure of heat production (calorimetry).
End organ damage is severe impairment of major body organs due to systemic disease. Commonly this is referred to in diabetes, high blood pressure, or states of low blood pressure or low blood volume. This can present as a heart attack or heart failure, pulmonary edema, neurologic deficits including a stroke, or acute kidney failure.
Starvation response in animals is a set of adaptive biochemical and physiological changes, triggered by lack of food or extreme weight loss, in which the body seeks to conserve energy by reducing metabolic rate and/or non-resting energy expenditure to prolong survival and preserve body fat and lean mass.
In molecular biology, the sulfonylurea receptors (SUR) are membrane proteins which are the molecular targets of the sulfonylurea class of antidiabetic drugs whose mechanism of action is to promote insulin release from pancreatic beta cells. More specifically, SUR proteins are subunits of the inward-rectifier potassium ion channels Kir6.x. The association of four Kir6.x and four SUR subunits form an ion conducting channel commonly referred to as the KATP channel.
Noninvasive glucose monitoring (NIGM), called Noninvasive continuous glucose monitoring when used as a CGM technique, is the measurement of blood glucose levels, required by people with diabetes to prevent both chronic and acute complications from the disease, without drawing blood, puncturing the skin, or causing pain or trauma. The search for a successful technique began about 1975 and has continued to the present without a clinically or commercially viable product.

In medicine, monitoring is the observation of a disease, condition or one or several medical parameters over time.
Fish are exposed to large oxygen fluctuations in their aquatic environment since the inherent properties of water can result in marked spatial and temporal differences in the concentration of oxygen. Fish respond to hypoxia with varied behavioral, physiological, and cellular responses to maintain homeostasis and organism function in an oxygen-depleted environment. The biggest challenge fish face when exposed to low oxygen conditions is maintaining metabolic energy balance, as 95% of the oxygen consumed by fish is used for ATP production releasing the chemical energy of nutrients through the mitochondrial electron transport chain. Therefore, hypoxia survival requires a coordinated response to secure more oxygen from the depleted environment and counteract the metabolic consequences of decreased ATP production at the mitochondria.
An organ-on-a-chip (OOC) is a multi-channel 3-D microfluidic cell culture, integrated circuit (chip) that simulates the activities, mechanics and physiological response of an entire organ or an organ system. It constitutes the subject matter of significant biomedical engineering research, more precisely in bio-MEMS. The convergence of labs-on-chips (LOCs) and cell biology has permitted the study of human physiology in an organ-specific context. By acting as a more sophisticated in vitro approximation of complex tissues than standard cell culture, they provide the potential as an alternative to animal models for drug development and toxin testing.
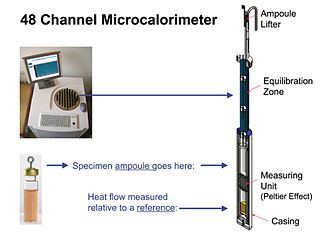
Isothermal microcalorimetry (IMC) is a laboratory method for real-time monitoring and dynamic analysis of chemical, physical and biological processes. Over a period of hours or days, IMC determines the onset, rate, extent and energetics of such processes for specimens in small ampoules at a constant set temperature.
In vitro models for calcification may refer to systems that have been developed in order to reproduce, in the best possible way, the calcification process that tissues or biomaterials undergo inside the body. The aim of these systems is to mimic the high levels of calcium and phosphate present in the blood and measure the extent of the crystal's deposition. Different variations can include other parameters to increase the veracity of these models, such as flow, pressure, compliance and resistance. All the systems have different limitations that have to be acknowledged regarding the operating conditions and the degree of representation. The rational of using such is to partially replace in vivo animal testing, whilst rendering much more controllable and independent parameters compared to an animal model.

A continuous glucose monitor (CGM) is a device used for monitoring blood glucose on a continual basis instead of monitoring glucose levels periodically by drawing a drop of blood from a finger. This is known as continuous glucose monitoring. CGMs are used by people who treat their diabetes with insulin, for example people with type 1 diabetes, type 2 diabetes, or other types of diabetes, such as gestational diabetes.
Bioinstrumentation or biomedical instrumentation is an application of biomedical engineering which focuses on development of devices and mechanics used to measure, evaluate, and treat biological systems. The goal of biomedical instrumentation focuses on the use of multiple sensors to monitor physiological characteristics of a human or animal for diagnostic and disease treatment purposes. Such instrumentation originated as a necessity to constantly monitor vital signs of Astronauts during NASA's Mercury, Gemini, and Apollo missions.
References
- ↑ McConnel HM, Owicki JC, Parce JW, Miller DL, Baxter GT, Wada HG, Pitchford S (1992). "The Cytosensor Microphysiometer: Biological Applications of Silicon Technology", Science, 257, 1906-1912
- ↑ Brischwein, M.; Wiest, J. (2018). "Microphysiometry". Bioanalytical Reviews. 2. Springer: 163–188. doi:10.1007/11663_2018_2. ISBN 978-3-030-32432-2.
- ↑ Hafeman DG, Parce JW, McConnell H (1988). "Light-addressable potentiometric sensor for biochemical systems", Science 240, 1182–1185
- ↑ Owicki JC, Parce JW (1990). "Bioassays with a microphysiometer". Nature 344, 271–272
- ↑ Wiest, J. (2022). "Systems engineering of microphysiometry". Organs-on-a-Chip. 4. Elsevier B.V. doi: 10.1016/j.ooc.2022.100016 .
- ↑ Alajoki ML, Bayter GT, Bemiss WR, Blau D, Bousse LJ, Chan SDH, Dawes TD, Hahnenberger KM, Hamilton JM, Lam P, McReynolds RJ, Modlin DN, Owicki C, Parce JW, Redington D, Stevenson K, Wada HG, Williams J (1997). "High-performance microphysiometry in drug discovery", Devlin JP (ed) High Throughput Screening: The Discovery of Bioactive Substances. Marcel Dekker, New York, 427–442.