Related Research Articles

Drug design, often referred to as rational drug design or simply rational design, is the inventive process of finding new medications based on the knowledge of a biological target. The drug is most commonly an organic small molecule that activates or inhibits the function of a biomolecule such as a protein, which in turn results in a therapeutic benefit to the patient. In the most basic sense, drug design involves the design of molecules that are complementary in shape and charge to the biomolecular target with which they interact and therefore will bind to it. Drug design frequently but not necessarily relies on computer modeling techniques. This type of modeling is sometimes referred to as computer-aided drug design. Finally, drug design that relies on the knowledge of the three-dimensional structure of the biomolecular target is known as structure-based drug design. In addition to small molecules, biopharmaceuticals including peptides and especially therapeutic antibodies are an increasingly important class of drugs and computational methods for improving the affinity, selectivity, and stability of these protein-based therapeutics have also been developed.
In the physical sciences, a partition coefficient (P) or distribution coefficient (D) is the ratio of concentrations of a compound in a mixture of two immiscible solvents at equilibrium. This ratio is therefore a comparison of the solubilities of the solute in these two liquids. The partition coefficient generally refers to the concentration ratio of un-ionized species of compound, whereas the distribution coefficient refers to the concentration ratio of all species of the compound.
Quantitative structure–activity relationship models are regression or classification models used in the chemical and biological sciences and engineering. Like other regression models, QSAR regression models relate a set of "predictor" variables (X) to the potency of the response variable (Y), while classification QSAR models relate the predictor variables to a categorical value of the response variable.

ADME is an abbreviation in pharmacokinetics and pharmacology for "absorption, distribution, metabolism, and excretion", and describes the disposition of a pharmaceutical compound within an organism. The four criteria all influence the drug levels and kinetics of drug exposure to the tissues and hence influence the performance and pharmacological activity of the compound as a drug. Sometimes, liberation and/or toxicity are also considered, yielding LADME, ADMET, or LADMET.
Cytochrome P450 3A4 is an important enzyme in the body, mainly found in the liver and in the intestine. It oxidizes small foreign organic molecules (xenobiotics), such as toxins or drugs, so that they can be removed from the body. It is highly homologous to CYP3A5, another important CYP3A enzyme.

Cytochrome P450 2E1 is a member of the cytochrome P450 mixed-function oxidase system, which is involved in the metabolism of xenobiotics in the body. This class of enzymes is divided up into a number of subcategories, including CYP1, CYP2, and CYP3, which as a group are largely responsible for the breakdown of foreign compounds in mammals.
Drug metabolism is the metabolic breakdown of drugs by living organisms, usually through specialized enzymatic systems. More generally, xenobiotic metabolism is the set of metabolic pathways that modify the chemical structure of xenobiotics, which are compounds foreign to an organism's normal biochemistry, such as any drug or poison. These pathways are a form of biotransformation present in all major groups of organisms and are considered to be of ancient origin. These reactions often act to detoxify poisonous compounds. The study of drug metabolism is called pharmacokinetics.
A toxicophore is a chemical structure or a portion of a structure that is related to the toxic properties of a chemical. Toxicophores can act directly or can require metabolic activation.

Cytochrome P450 family 2 subfamily C member 9 is an enzyme protein. The enzyme is involved in metabolism, by oxidation, of both xenobiotics, including drugs, and endogenous compounds, including fatty acids. In humans, the protein is encoded by the CYP2C9 gene. The gene is highly polymorphic, which affects the efficiency of the metabolism by the enzyme.
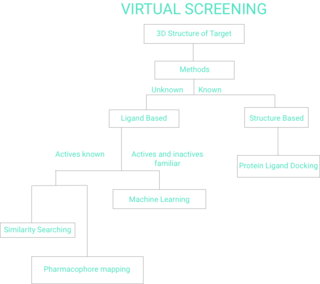
Virtual screening (VS) is a computational technique used in drug discovery to search libraries of small molecules in order to identify those structures which are most likely to bind to a drug target, typically a protein receptor or enzyme.
Hit to lead (H2L) also known as lead generation is a stage in early drug discovery where small molecule hits from a high throughput screen (HTS) are evaluated and undergo limited optimization to identify promising lead compounds. These lead compounds undergo more extensive optimization in a subsequent step of drug discovery called lead optimization (LO). The drug discovery process generally follows the following path that includes a hit to lead stage:
Pharmacokinetics, sometimes abbreviated as PK, is a branch of pharmacology dedicated to determining the fate of substances administered to a living organism. The substances of interest include any chemical xenobiotic such as: pharmaceutical drugs, pesticides, food additives, cosmetics, etc. It attempts to analyze chemical metabolism and to discover the fate of a chemical from the moment that it is administered up to the point at which it is completely eliminated from the body. Pharmacokinetics is the study of how an organism affects a drug, whereas pharmacodynamics (PD) is the study of how the drug affects the organism. Both together influence dosing, benefit, and adverse effects, as seen in PK/PD models.
Integrated discrete Multiple Organ Culture (IdMOC) is an in vitro, cell culture based experimental model for the study of intercellular communication. In conventional in vitro systems, each cell type is studied in isolation ignoring critical interactions between organs or cell types. IdMOC technology is based on the concept that multiple organs signal or communicate via the systemic circulation (i.e., blood).
Simcyp Limited is a research-based company which provides modelling and simulation software to the pharmaceutical industry for use during drug development. Simcyp is based in Sheffield, UK.
Inte:Ligand was founded in Maria Enzersdorf, Lower Austria (Niederösterreich) in 2003. They established the company headquarters on Mariahilferstrasse in Vienna, Austria that same year.
Computational Resources for Drug Discovery (CRDD) is one of the important silico modules of Open Source for Drug Discovery (OSDD). The CRDD web portal provides computer resources related to drug discovery on a single platform. It provides computational resources for researchers in computer-aided drug design, a discussion forum, and resources to maintain Wikipedia related to drug discovery, predict inhibitors, and predict the ADME-Tox property of molecules One of the major objectives of CRDD is to promote open source software in the field of chemoinformatics and pharmacoinformatics.
Advanced Chemistry Development, Inc., (ACD/Labs) specializes in the design of software, with a focus on the R&D and chemistry of molecules. ACD/Labs provides solutions for the enterprise in varied areas, including analytical data handling and knowledge management; in addition, molecular property modelling and property-based design are relevant areas of expertise in ACD Labs.
Discovery Studio is a suite of software for simulating small molecule and macromolecule systems. It is developed and distributed by Dassault Systemes BIOVIA.

Sean Ekins is a British pharmacologist and expert in the fields of ADME/Tox, computational toxicology and cheminformatics at Collaborations in Chemistry, a division of corporate communications firm Collaborations in Communications. He is also the editor of four books and a book series for John Wiley & Sons.
Pharmaceutical bioinformatics is a research field related to bioinformatics but with the focus on studying biological and chemical processes in the pharmaceutical area; to understand how xenobiotics interact with the human body and the drug discovery process.
References
- ↑ Goodford, P.J. (1985) A computational procedure for determining energetically favorable binding sites on biologically important macromolecules. J. Med. Chem., 28, 849-857
- ↑ Von Itzstein, M. et al. (1993) Rational design of potent sialidase-based inhibitors of influenza virus replication. Nature, 363, 418-423
- ↑ Cytochrome P450 Consortium