Oxidation of ammonia to nitrate
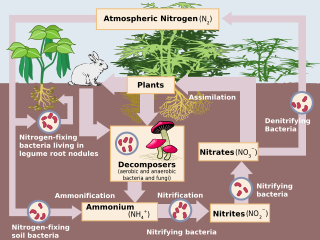
Nitrification in nature is a two-step oxidation process of ammonium (NH+4) or ammonia (NH3) to nitrite (NO−2) and then to nitrate (NO−3) catalyzed by two ubiquitous bacterial groups growing together. The first reaction is oxidation of ammonium to nitrite by ammonia oxidizing bacteria (AOB) represented by members of Betaproteobacteria and Gammaproteobacteria. Further organisms able to oxidize ammonia are Archaea (AOA). [4]
The second reaction is oxidation of nitrite (NO−2) to nitrate by nitrite-oxidizing bacteria (NOB), represented by the members of Nitrospinota, Nitrospirota, Pseudomonadota, and Chloroflexota. [5] [6]
This two-step process was described already in 1890 by the Ukrainian microbiologist Sergei Winogradsky.
Ammonia can be also oxidized completely to nitrate by one comammox bacterium.
Ammonia-to-nitrite mechanism

Ammonia oxidation in autotrophic nitrification is a complex process that requires several enzymes as well as oxygen as a reactant. The key enzymes necessary for releasing energy during oxidation of ammonia to nitrite are ammonia monooxygenase (AMO) and hydroxylamine oxidoreductase (HAO). The first is a transmembrane copper protein which catalyzes the oxidation of ammonia to hydroxylamine ( 1.1 ) taking two electrons directly from the quinone pool. This reaction requires O2.
The second step of this process has recently fallen into question. [7] For the past few decades, the common view was that a trimeric multiheme c-type HAO converts hydroxylamine into nitrite in the periplasm with production of four electrons ( 1.2 ). The stream of four electrons is channeled through cytochrome c554 to a membrane-bound cytochrome c552. Two of the electrons are routed back to AMO, where they are used for the oxidation of ammonia (quinol pool). The remaining two electrons are used to generate a proton motive force and reduce NAD(P) through reverse electron transport. [8]
Recent results, however, show that HAO does not produce nitrite as a direct product of catalysis. This enzyme instead produces nitric oxide and three electrons. Nitric oxide can then be oxidized by other enzymes (or oxygen) to nitrite. In this paradigm, the electron balance for overall metabolism needs to be reconsidered. [7]
- NH3 + O2 → NO−2 + 3H+ + 2e−
(1)
- NH3 + O2 + 2H+ + 2e− → NH2OH + H2O
(1.1)
- NH2OH + H2O → NO−2 + 5H+ + 4e−
(1.2)
Nitrite-to-nitrate mechanism
Nitrite produced in the first step of autotrophic nitrification is oxidized to nitrate by nitrite oxidoreductase (NXR) ( 2 ). It is a membrane-associated iron-sulfur molybdo protein and is part of an electron transfer chain which channels electrons from nitrite to molecular oxygen.[ citation needed ] The enzymatic mechanisms involved in nitrite-oxidizing bacteria are less described than that of ammonium oxidation. Recent research (e.g. Woźnica A. et al., 2013) [9] proposes a new hypothetical model of NOB electron transport chain and NXR mechanisms. Here, in contrast to earlier models, [10] the NXR would act on the outside of the plasma membrane and directly contribute to a mechanism of proton gradient generation as postulated by Spieck [11] and coworkers. Nevertheless, the molecular mechanism of nitrite oxidation is an open question.
(2)
Comammox bacteria
The two-step conversion of ammonia to nitrate observed in ammonia-oxidizing bacteria, ammonia-oxidizing archaea and nitrite-oxidizing bacteria (such as Nitrobacter) is puzzling to researchers. [12] [13] Complete nitrification, the conversion of ammonia to nitrate in a single step known as comammox, has an energy yield (∆G°′) of −349 kJ mol−1 NH3, while the energy yields for the ammonia-oxidation and nitrite-oxidation steps of the observed two-step reaction are −275 kJ mol−1 NH3, and −74 kJ mol−1 NO2−, respectively. [12] These values indicate that it would be energetically favourable for an organism to carry out complete nitrification from ammonia to nitrate (comammox), rather than conduct only one of the two steps. The evolutionary motivation for a decoupled, two-step nitrification reaction is an area of ongoing research. In 2015, it was discovered that the species Nitrospira inopinata possesses all the enzymes required for carrying out complete nitrification in one step, suggesting that this reaction does occur. [12] [13]
Table of characteristics
| Genus | Phylogenetic group | DNA (mol% GC) | Habitats | Characteristics |
|---|---|---|---|---|
Nitrifying bacteria that oxidize ammonia [5] [14] | ||||
| Nitrosomonas | Beta | 45-53 | Soil, sewage, freshwater, marine | Gram-negative short to long rods, motile (polar flagella) or nonmotile; peripheral membrane systems |
| Nitrosococcus | Gamma | 49-50 | Freshwater, marine | Large cocci, motile, vesicular or peripheral membranes |
| Nitrosospira | Beta | 54 | Soil | Spirals, motile (peritrichous flagella); no obvious membrane system |
Nitrifying bacteria that oxidize nitrite [5] [14] | ||||
| Nitrobacter | Alpha | 59-62 | Soil, freshwater, marine | Short rods, reproduce by budding, occasionally motile (single subterminal flagella) or non-motile; membrane system arranged as a polar cap |
| Nitrospina | Delta | 58 | Marine | Long, slender rods, nonmotile, no obvious membrane system |
| Nitrococcus | Gamma | 61 | Marine | Large cocci, motile (one or two subterminal flagellum) membrane system randomly arranged in tubes |
| Nitrospira | Nitrospirota | 50 | Marine, soil | Helical to vibroid-shaped cells; nonmotile; no internal membranes |
Comammox bacteria [15] [16] [17] | ||||
| Nitrospira inopinata | Nitrospirota | 59.23 | Microbial mat in hot water pipes (56 °C, pH 7.5) | Rods |