Related Research Articles

Titration is a common laboratory method of quantitative chemical analysis to determine the concentration of an identified analyte. A reagent, termed the titrant or titrator, is prepared as a standard solution of known concentration and volume. The titrant reacts with a solution of analyte to determine the analyte's concentration. The volume of titrant that reacted with the analyte is termed the titration volume.
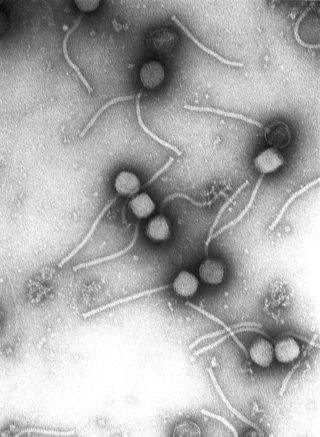
Virology is the scientific study of biological viruses. It is a subfield of microbiology that focuses on their detection, structure, classification and evolution, their methods of infection and exploitation of host cells for reproduction, their interaction with host organism physiology and immunity, the diseases they cause, the techniques to isolate and culture them, and their use in research and therapy.

Centrifugation is a mechanical process which involves the use of the centrifugal force to separate particles from a solution according to their size, shape, density, medium viscosity and rotor speed. The denser components of the mixture migrate away from the axis of the centrifuge, while the less dense components of the mixture migrate towards the axis. Chemists and biologists may increase the effective gravitational force of the test tube so that the precipitate (pellet) will travel quickly and fully to the bottom of the tube. The remaining liquid that lies above the precipitate is called a supernatant or supernate.
An assay is an investigative (analytic) procedure in laboratory medicine, mining, pharmacology, environmental biology and molecular biology for qualitatively assessing or quantitatively measuring the presence, amount, or functional activity of a target entity. The measured entity is often called the analyte, the measurand, or the target of the assay. The analyte can be a drug, biochemical substance, chemical element or compound, or cell in an organism or organic sample. An assay usually aims to measure an analyte's intensive property and express it in the relevant measurement unit.
Protein purification is a series of processes intended to isolate one or a few proteins from a complex mixture, usually cells, tissues or whole organisms. Protein purification is vital for the specification of the function, structure and interactions of the protein of interest. The purification process may separate the protein and non-protein parts of the mixture, and finally separate the desired protein from all other proteins. Ideally, to study a protein of interest, it must be separated from other components of the cell so that contaminants won't interfere in the examination of the protein of interest's structure and function. Separation of one protein from all others is typically the most laborious aspect of protein purification. Separation steps usually exploit differences in protein size, physico-chemical properties, binding affinity and biological activity. The pure result may be termed protein isolate.
Plate readers, also known as microplate readers or microplate photometers, are instruments which are used to detect biological, chemical or physical events of samples in microtiter plates. They are widely used in research, drug discovery, bioassay validation, quality control and manufacturing processes in the pharmaceutical and biotechnological industry and academic organizations. Sample reactions can be assayed in 1-1536 well format microtiter plates. The most common microplate format used in academic research laboratories or clinical diagnostic laboratories is 96-well with a typical reaction volume between 100 and 200 µL per well. Higher density microplates are typically used for screening applications, when throughput and assay cost per sample become critical parameters, with a typical assay volume between 5 and 50 µL per well. Common detection modes for microplate assays are absorbance, fluorescence intensity, luminescence, time-resolved fluorescence, and fluorescence polarization.
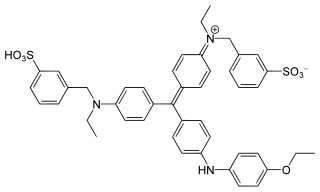
Coomassie brilliant blue is the name of two similar triphenylmethane dyes that were developed for use in the textile industry but are now commonly used for staining proteins in analytical biochemistry. Coomassie brilliant blue G-250 differs from Coomassie brilliant blue R-250 by the addition of two methyl groups. The name "Coomassie" is a registered trademark of Imperial Chemical Industries.
Ammonium sulfate precipitation is one of the most commonly used methods for large and laboratory scale protein purification and fractionation that can be used to separate proteins by altering their solubility in the presence of a high salt concentration.
The Bradford protein assay was developed by Marion M. Bradford in 1976. It is a quick and accurate spectroscopic analytical procedure used to measure the concentration of protein in a solution. The reaction is dependent on the amino acid composition of the measured proteins.

Saponification value or saponification number represents the number of milligrams of potassium hydroxide (KOH) or sodium hydroxide (NaOH) required to saponify one gram of fat under the conditions specified. It is a measure of the average molecular weight of all the fatty acids present in the sample in form of triglycerides. The higher the saponification value, the lower the fatty acids average length, the lighter the mean molecular weight of triglycerides and vice versa. Practically, fats or oils with high saponification value are more suitable for soap making.
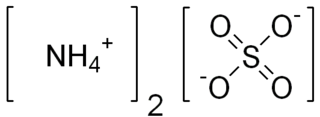
Ammonium sulfate (American English and international scientific usage; ammonium sulphate in British English); (NH4)2SO4, is an inorganic salt with a number of commercial uses. The most common use is as a soil fertilizer. It contains 21% nitrogen and 24% sulfur.
Protein methods are the techniques used to study proteins. There are experimental methods for studying proteins. Computational methods typically use computer programs to analyze proteins. However, many experimental methods require computational analysis of the raw data.

In chemistry, the Biuret test, also known as Piotrowski's test, is a chemical test used for detecting the presence of at least two peptide bonds in a molecule. In the presence of peptides, a copper(II) ion forms mauve-colored coordination complexes in an alkaline solution. The reaction was first observed in 1833; In Poland, the biuret test is also known as Piotrowski's test in honor of the Polish physiologist Gustaw Piotrowski who independently rediscovered it in 1857. Several variants on the test have been developed, such as the BCA test and the Modified Lowry test.
The Kjeldahl method or Kjeldahl digestion (Danish pronunciation: [ˈkʰelˌtɛˀl]) in analytical chemistry is a method for the quantitative determination of nitrogen contained in organic substances plus the nitrogen contained in the inorganic compounds ammonia and ammonium (NH3/NH4+). Without modification, other forms of inorganic nitrogen, for instance nitrate, are not included in this measurement. Using an empirical relation between Kjeldahl nitrogen content and protein content it is an important method for analyzing proteins. This method was developed by Johan Kjeldahl in 1883.

Soy protein is a protein that is isolated from soybean. It is made from soybean meal that has been dehulled and defatted. Dehulled and defatted soybeans are processed into three kinds of high protein commercial products: soy flour, concentrates, and isolates. Soy protein isolate has been used since 1959 in foods for its functional properties.

The Folin–Ciocâlteu reagent (FCR) or Folin's phenol reagent or Folin–Denis reagent, is a mixture of phosphomolybdate and phosphotungstate used for the colorimetric in vitro assay of phenolic and polyphenolic antioxidants, also called the gallic acid equivalence method (GAE). It is named after Otto Folin, Vintilă Ciocâlteu, and Willey Glover Denis. The Folin-Denis reagent is prepared by mixing sodium tungstate and phosphomolybdic acid in phosphoric acid. The Folin–Ciocalteu reagent is just a modification of the Folin-Denis reagent. The modification consisted of the addition of lithium sulfate and bromine to the phosphotungstic-phosphomolybdic reagent.
The Gerber method is a primary and historic chemical test to determine the fat content of substances, most commonly milk and cream. The Gerber method is the primary testing method in Europe and much of the world. The fairly similar Babcock test is used primarily in the United States, although the Gerber method also enjoys significant use in the U.S. as well.
The Dumas method in analytical chemistry is a method for the quantitative determination of nitrogen in chemical substances based on a method first described by Jean-Baptiste Dumas in 1826.
A thermal shift assay (TSA) measures changes in the thermal denaturation temperature and hence stability of a protein under varying conditions such as variations in drug concentration, buffer pH or ionic strength, redox potential, or sequence mutation. The most common method for measuring protein thermal shifts is differential scanning fluorimetry (DSF) or thermofluor, which utilizes specialized fluorogenic dyes.
A ligand binding assay (LBA) is an assay, or an analytic procedure, which relies on the binding of ligand molecules to receptors, antibodies or other macromolecules. A detection method is used to determine the presence and extent of the ligand-receptor complexes formed, and this is usually determined electrochemically or through a fluorescence detection method. This type of analytic test can be used to test for the presence of target molecules in a sample that are known to bind to the receptor.
References
- ↑ Zayas, Joseph F. (2012-12-06). Functionality of Proteins in Food. Springer Science & Business Media. ISBN 978-3-642-59116-7.
- ↑ Nollet, Leo M. L.; Toldra, Fidel (2009-11-04). Handbook of Dairy Foods Analysis. CRC Press. ISBN 978-1-4200-4631-1.
- ↑ Damodaran, Srinivasan (2017-10-19). Food Proteins and Their Applications. Routledge. ISBN 978-1-351-44752-2.
- ↑ Nielsen, Suzanne (2003-04-30). Food Analysis. Springer Science & Business Media. ISBN 978-0-306-47495-8.
- ↑ Hall, G. M. (1996-06-30). Methods of Testing Protein Functionality. Springer Science & Business Media. ISBN 978-0-7514-0053-3.