Related Research Articles

A zygote is a eukaryotic cell formed by a fertilization event between two gametes. The zygote's genome is a combination of the DNA in each gamete, and contains all of the genetic information of a new individual organism.

Amniocentesis is a medical procedure used primarily in the prenatal diagnosis of genetic conditions. It has other uses such as in the assessment of infection and fetal lung maturity. Prenatal diagnostic testing, which includes amniocentesis, is necessary to conclusively diagnose the majority of genetic disorders, with amniocentesis being the gold-standard procedure after 15 weeks' gestation.

Aneuploidy is the presence of an abnormal number of chromosomes in a cell, for example a human cell having 45 or 47 chromosomes instead of the usual 46. It does not include a difference of one or more complete sets of chromosomes. A cell with any number of complete chromosome sets is called a euploid cell.

Genetic testing, also known as DNA testing, is used to identify changes in DNA sequence or chromosome structure. Genetic testing can also include measuring the results of genetic changes, such as RNA analysis as an output of gene expression, or through biochemical analysis to measure specific protein output. In a medical setting, genetic testing can be used to diagnose or rule out suspected genetic disorders, predict risks for specific conditions, or gain information that can be used to customize medical treatments based on an individual's genetic makeup. Genetic testing can also be used to determine biological relatives, such as a child's biological parentage through DNA paternity testing, or be used to broadly predict an individual's ancestry. Genetic testing of plants and animals can be used for similar reasons as in humans, to gain information used for selective breeding, or for efforts to boost genetic diversity in endangered populations.
Prenatal testing consists of prenatal screening and prenatal diagnosis, which are aspects of prenatal care that focus on detecting problems with the pregnancy as early as possible. These may be anatomic and physiologic problems with the health of the zygote, embryo, or fetus, either before gestation even starts or as early in gestation as practicable. Screening can detect problems such as neural tube defects, chromosome abnormalities, and gene mutations that would lead to genetic disorders and birth defects, such as spina bifida, cleft palate, Down syndrome, Tay–Sachs disease, sickle cell anemia, thalassemia, cystic fibrosis, muscular dystrophy, and fragile X syndrome. Some tests are designed to discover problems which primarily affect the health of the mother, such as PAPP-A to detect pre-eclampsia or glucose tolerance tests to diagnose gestational diabetes. Screening can also detect anatomical defects such as hydrocephalus, anencephaly, heart defects, and amniotic band syndrome.

Chorionic villus sampling (CVS), sometimes called "chorionic villous sampling", is a form of prenatal diagnosis done to determine chromosomal or genetic disorders in the fetus. It entails sampling of the chorionic villus and testing it for chromosomal abnormalities, usually with FISH or PCR. CVS usually takes place at 10–12 weeks' gestation, earlier than amniocentesis or percutaneous umbilical cord blood sampling. It is the preferred technique before 26 days.
Echogenic intracardiac focus (EIF) is a small bright spot seen in the baby's heart on an ultrasound exam. This is thought to represent mineralization, or small deposits of calcium, in the muscle of the heart. EIFs are found in about 3–5% of normal pregnancies and cause no health problems.

Genetic analysis is the overall process of studying and researching in fields of science that involve genetics and molecular biology. There are a number of applications that are developed from this research, and these are also considered parts of the process. The base system of analysis revolves around general genetics. Basic studies include identification of genes and inherited disorders. This research has been conducted for centuries on both a large-scale physical observation basis and on a more microscopic scale. Genetic analysis can be used generally to describe methods both used in and resulting from the sciences of genetics and molecular biology, or to applications resulting from this research.
A genomic library is a collection of overlapping DNA fragments that together make up the total genomic DNA of a single organism. The DNA is stored in a population of identical vectors, each containing a different insert of DNA. In order to construct a genomic library, the organism's DNA is extracted from cells and then digested with a restriction enzyme to cut the DNA into fragments of a specific size. The fragments are then inserted into the vector using DNA ligase. Next, the vector DNA can be taken up by a host organism - commonly a population of Escherichia coli or yeast - with each cell containing only one vector molecule. Using a host cell to carry the vector allows for easy amplification and retrieval of specific clones from the library for analysis.
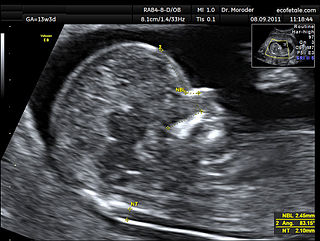
A nuchal scan or nuchal translucency (NT) scan/procedure is a sonographic prenatal screening scan (ultrasound) to detect chromosomal abnormalities in a fetus, though altered extracellular matrix composition and limited lymphatic drainage can also be detected.

Molecular cytogenetics combines two disciplines, molecular biology and cytogenetics, and involves the analysis of chromosome structure to help distinguish normal and cancer-causing cells. Human cytogenetics began in 1956 when it was discovered that normal human cells contain 46 chromosomes. However, the first microscopic observations of chromosomes were reported by Arnold, Flemming, and Hansemann in the late 1800s. Their work was ignored for decades until the actual chromosome number in humans was discovered as 46. In 1879, Arnold examined sarcoma and carcinoma cells having very large nuclei. Today, the study of molecular cytogenetics can be useful in diagnosing and treating various malignancies such as hematological malignancies, brain tumors, and other precursors of cancer. The field is overall focused on studying the evolution of chromosomes, more specifically the number, structure, function, and origin of chromosome abnormalities. It includes a series of techniques referred to as fluorescence in situ hybridization, or FISH, in which DNA probes are labeled with different colored fluorescent tags to visualize one or more specific regions of the genome. Introduced in the 1980s, FISH uses probes with complementary base sequences to locate the presence or absence of the specific DNA regions. FISH can either be performed as a direct approach to metaphase chromosomes or interphase nuclei. Alternatively, an indirect approach can be taken in which the entire genome can be assessed for copy number changes using virtual karyotyping. Virtual karyotypes are generated from arrays made of thousands to millions of probes, and computational tools are used to recreate the genome in silico.

Trisomy 16 is a chromosomal abnormality in which there are 3 copies of chromosome 16 rather than two. It is the most common trisomy leading to miscarriage and the second most common chromosomal cause of it, closely following X-chromosome monosomy. About 6% of miscarriages have trisomy 16. Those mostly occur between 8 and 15 weeks after the last menstrual period.

Whole genome sequencing (WGS), also known as full genome sequencing, complete genome sequencing, or entire genome sequencing, is the process of determining the entirety, or nearly the entirety, of the DNA sequence of an organism's genome at a single time. This entails sequencing all of an organism's chromosomal DNA as well as DNA contained in the mitochondria and, for plants, in the chloroplast.
Sequenom is an American company based in San Diego, California. It develops enabling molecular technologies, and highly sensitive laboratory genetic tests for NIPT. Sequenom's wholly owned subsidiarity, Sequenom Center for Molecular Medicine (SCMM), offers multiple clinical molecular genetics tests to patients, including MaterniT21, plus a noninvasive prenatal test for trisomy 21, trisomy 18, and trisomy 13, and the SensiGene RHD Fetal RHD genotyping test.
Ravinder (Rav) Dhallan is the chairman and chief executive officer of Ravgen.
Natera, Inc. is a clinical genetic testing company based in Austin, Texas that specializes in non-invasive, cell-free DNA (cfDNA) testing technology, with a focus on women’s health, cancer, and organ health. Natera’s proprietary technology combines novel molecular biology techniques with a suite of bioinformatics software that allows detection down to a single molecule in a tube of blood. Natera operates CAP-accredited laboratories certified under the Clinical Laboratory Improvement Amendments (CLIA) in San Carlos, California and Austin, Texas.
Cell-free fetal DNA (cffDNA) is fetal DNA that circulates freely in the maternal blood. Maternal blood is sampled by venipuncture. Analysis of cffDNA is a method of non-invasive prenatal diagnosis frequently ordered for pregnant women of advanced maternal age. Two hours after delivery, cffDNA is no longer detectable in maternal blood.
Dennis Yuk Ming Lo is a Hong Kong molecular biologist, and an important contributor to the development of non-invasive prenatal testing. He is the current Associate Dean (Research) and Li Ka Shing Professor of Medicine at the Chinese University of Hong Kong (CUHK), as well as the head of the Department of Chemical Pathology of CUHK and the director of the Li Ka Shing Institute of Health Sciences. His research focuses on the detection of cell-free fetal DNA in blood plasma.

Diana W. Bianchi is an American medical geneticist and neonatologist noted for her research on fetal cell microchimerism and prenatal testing. She is the director of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, part of the National Institutes of Health. Bianchi had previously been the Natalie V. Zucker Professor of Pediatrics, Obstetrics, and Gynecology at Tufts University School of Medicine and founder and executive director of the Mother Infant Research Institute at Tufts Medical Center. She also has served as Vice Chair for Research in the Department of Pediatrics at the Floating Hospital for Children at Tufts Medical Center.
Noninvasive prenatal testing (NIPT) is a method used to determine the risk for the fetus being born with certain chromosomal abnormalities, such as trisomy 21, trisomy 18 and trisomy 13. This testing analyzes small DNA fragments that circulate in the blood of a pregnant woman. Unlike most DNA found in the nucleus of a cell, these fragments are not found within the cells, instead they are free-floating, and so are called cell free fetal DNA (cffDNA). These fragments usually contain less than 200 DNA building blocks (base pairs) and arise when cells die, and their contents, including DNA, are released into the bloodstream. cffDNA derives from placental cells and is usually identical to fetal DNA. Analysis of cffDNA from placenta provides the opportunity for early detection of certain chromosomal abnormalities without harming the fetus.
References
- ↑ "Clinical Data". Verinata. 2012-01-11. Retrieved 2013-08-19.
- ↑ Sehnert AJ, et al. (Jul 2011). "Optimal detection of fetal chromosomal abnormalities by massively parallel DNA sequencing of cell-free fetal DNA from maternal blood". Clin. Chem. 57 (7): 1042–9. doi: 10.1373/clinchem.2011.165910 . PMID 21519036.