Related Research Articles

In genetics, a promoter is a sequence of DNA to which proteins bind that initiate transcription of a single RNA from the DNA downstream of it. This RNA may encode a protein, or can have a function in and of itself, such as tRNA, mRNA, or rRNA. Promoters are located near the transcription start sites of genes, upstream on the DNA . Promoters can be about 100–1000 base pairs long.

Transcription is the first of several steps of DNA based gene expression in which a particular segment of DNA is copied into RNA by the enzyme RNA polymerase.

In molecular biology, RNA polymerase, is an enzyme that synthesizes RNA from a DNA template.
In molecular biology and genetics, transcriptional regulation is the means by which a cell regulates the conversion of DNA to RNA (transcription), thereby orchestrating gene activity. A single gene can be regulated in a range of ways, from altering the number of copies of RNA that are transcribed, to the temporal control of when the gene is transcribed. This control allows the cell or organism to respond to a variety of intra- and extracellular signals and thus mount a response. Some examples of this include producing the mRNA that encode enzymes to adapt to a change in a food source, producing the gene products involved in cell cycle specific activities, and producing the gene products responsible for cellular differentiation in multicellular eukaryotes, as studied in evolutionary developmental biology.
A sigma factor is a protein needed for initiation of transcription in bacteria. It is a bacterial transcription initiation factor that enables specific binding of RNA polymerase (RNAP) to gene promoters. It is homologous to archaeal transcription factor B and to eukaryotic factor TFIIB. The specific sigma factor used to initiate transcription of a given gene will vary, depending on the gene and on the environmental signals needed to initiate transcription of that gene. Selection of promoters by RNA polymerase is dependent on the sigma factor that associates with it. They are also found in plant chloroplasts as a part of the bacteria-like plastid-encoded polymerase (PEP).
In molecular biology, the TATA box is a sequence of DNA found in the core promoter region of genes in archaea and eukaryotes. The bacterial homolog of the TATA box is called the Pribnow box which has a shorter consensus sequence.
A transcriptional activator is a protein that increases transcription of a gene or set of genes. Activators are considered to have positive control over gene expression, as they function to promote gene transcription and, in some cases, are required for the transcription of genes to occur. Most activators are DNA-binding proteins that bind to enhancers or promoter-proximal elements. The DNA site bound by the activator is referred to as an "activator-binding site". The part of the activator that makes protein–protein interactions with the general transcription machinery is referred to as an "activating region" or "activation domain".

General transcription factors (GTFs), also known as basal transcriptional factors, are a class of protein transcription factors that bind to specific sites (promoter) on DNA to activate transcription of genetic information from DNA to messenger RNA. GTFs, RNA polymerase, and the mediator constitute the basic transcriptional apparatus that first bind to the promoter, then start transcription. GTFs are also intimately involved in the process of gene regulation, and most are required for life.

Catabolite activator protein is a trans-acting transcriptional activator that exists as a homodimer in solution. Each subunit of CAP is composed of a ligand-binding domain at the N-terminus and a DNA-binding domain at the C-terminus. Two cAMP molecules bind dimeric CAP with negative cooperativity. Cyclic AMP functions as an allosteric effector by increasing CAP's affinity for DNA. CAP binds a DNA region upstream from the DNA binding site of RNA Polymerase. CAP activates transcription through protein-protein interactions with the α-subunit of RNA Polymerase. This protein-protein interaction is responsible for (i) catalyzing the formation of the RNAP-promoter closed complex; and (ii) isomerization of the RNAP-promoter complex to the open confirmation. CAP's interaction with RNA polymerase causes bending of the DNA near the transcription start site, thus effectively catalyzing the transcription initiation process. CAP's name is derived from its ability to affect transcription of genes involved in many catabolic pathways. For example, when the amount of glucose transported into the cell is low, a cascade of events results in the increase of cytosolic cAMP levels. This increase in cAMP levels is sensed by CAP, which goes on to activate the transcription of many other catabolic genes.
In eukaryote cells, RNA polymerase III transcribes DNA to synthesize ribosomal 5S rRNA, tRNA and other small RNAs.

The TATA-binding protein (TBP) is a general transcription factor that binds specifically to a DNA sequence called the TATA box. This DNA sequence is found about 30 base pairs upstream of the transcription start site in some eukaryotic gene promoters.
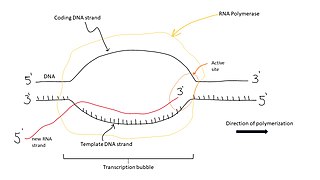
A transcription bubble is a molecular structure formed during DNA transcription when a limited portion of the DNA double strand is unwound. The size of a transcription bubble ranges from 12-14 base pairs. A transcription bubble is formed when the RNA polymerase enzyme binds to a promoter and causes two DNA strands to detach. It presents a region of unpaired DNA, where a short stretch of nucleotides are exposed on each strand of the double helix.
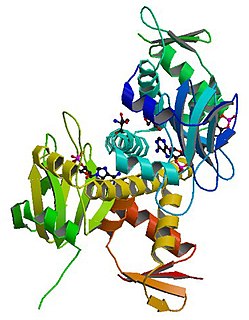
cAMP receptor protein is a regulatory protein in bacteria. CRP protein binds cAMP, which causes a conformational change that allows CRP to bind tightly to a specific DNA site in the promoters of the genes it controls. CRP then activates transcription through direct protein–protein interactions with RNA polymerase.

Bacterial transcription is the process in which a segment of bacterial DNA is copied into a newly synthesized strand of messenger RNA (mRNA) with use of the enzyme RNA polymerase. The process occurs in three main steps: initiation, elongation, and termination; and the end result is a strand of mRNA that is complementary to a single strand of DNA. Generally, the transcribed region accounts for more than one gene. In fact, many prokaryotic genes occur in operons, which are a series of genes that work together to code for the same protein or gene product and are controlled by a single promoter. Bacterial RNA polymerase is made up of four subunits and when a fifth subunit attaches, called the σ-factor, the polymerase can recognize specific binding sequences in the DNA, called promoters. The binding of the σ-factor to the promoter is the first step in initiation. Once the σ-factor releases from the polymerase, elongation proceeds. The polymerase continues down the double stranded DNA, unwinding it and synthesizing the new mRNA strand until it reaches a termination site. There are two termination mechanisms that are discussed in further detail below. Termination is required at specific sites for proper gene expression to occur. Gene expression determines how much gene product, such as protein, is made by the gene. Transcription is carried out by RNA polymerase but its specificity is controlled by sequence-specific DNA binding proteins called transcription factors. Transcription factors work to recognize specific DNA sequences and based on the cells needs, promote or inhibit additional transcription.

Eukaryotic transcription is the elaborate process that eukaryotic cells use to copy genetic information stored in DNA into units of transportable complementary RNA replica. Gene transcription occurs in both eukaryotic and prokaryotic cells. Unlike prokaryotic RNA polymerase that initiates the transcription of all different types of RNA, RNA polymerase in eukaryotes comes in three variations, each translating a different type of gene. A eukaryotic cell has a nucleus that separates the processes of transcription and translation. Eukaryotic transcription occurs within the nucleus where DNA is packaged into nucleosomes and higher order chromatin structures. The complexity of the eukaryotic genome necessitates a great variety and complexity of gene expression control.
Transcription factor TFIIA is a nuclear protein involved in the RNA polymerase II-dependent transcription of DNA. TFIIA is one of several general (basal) transcription factors (GTFs) that are required for all transcription events that use RNA polymerase II. Other GTFs include TFIID, a complex composed of the TATA binding protein TBP and TBP-associated factors (TAFs), as well as the factors TFIIB, TFIIE, TFIIF, and TFIIH. Together, these factors are responsible for promoter recognition and the formation of a transcription preinitiation complex (PIC) capable of initiating RNA synthesis from a DNA template.
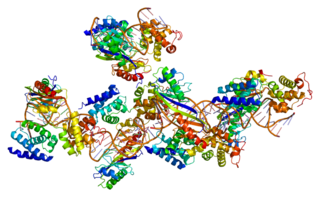
Transcription factor II B (TFIIB) is a general transcription factor that is involved in the formation of the RNA polymerase II preinitiation complex (PIC) and aids in stimulating transcription initiation. TFIIB is localised to the nucleus and provides a platform for PIC formation by binding and stabilising the DNA-TBP complex and by recruiting RNA polymerase II and other transcription factors. It is encoded by the TFIIB gene, and is homologous to archaeal transcription factor B and analogous to bacterial sigma factors.
The gal operon is a prokaryotic operon, which encodes enzymes necessary for galactose metabolism. Repression of gene expression for this operon works via binding of repressor molecules to two operators. These repressors dimerize, creating a loop in the DNA. The loop as well as hindrance from the external operator prevent RNA polymerase from binding to the promoter, and thus prevent transcription. Additionally, since the metabolism of galactose in the cell is involved in both anabolic and catabolic pathways, a novel regulatory system using two promoters for differential repression has been identified and characterized within the context of the gal operon.
RNA polymerase II holoenzyme is a form of eukaryotic RNA polymerase II that is recruited to the promoters of protein-coding genes in living cells. It consists of RNA polymerase II, a subset of general transcription factors, and regulatory proteins known as SRB proteins.
The gene rpoN encodes the sigma factor sigma-54, a protein in Escherichia coli and other species of bacteria. RpoN antagonizes RpoS sigma factors.
References
- ↑ Carmona, M; Magasanik, B (1996). "Activation of transcription at sigma 54-dependent promoters on linear templates requires intrinsic or induced bending of the DNA". J Mol Biol. 261 (3): 348–56. doi:10.1006/jmbi.1996.0468. PMID 8780778.
- Lecture notes, Molecular Genetics 3 at the University of Edinburgh autumn 2006