Related Research Articles

Cladistics is an approach to biological classification in which organisms are categorized in groups ("clades") based on hypotheses of most recent common ancestry. The evidence for hypothesized relationships is typically shared derived characteristics (synapomorphies) that are not present in more distant groups and ancestors. However, from an empirical perspective, common ancestors are inferences based on a cladistic hypothesis of relationships of taxa whose character states can be observed. Theoretically, a last common ancestor and all its descendants constitute a (minimal) clade. Importantly, all descendants stay in their overarching ancestral clade. For example, if the terms worms or fishes were used within a strict cladistic framework, these terms would include humans. Many of these terms are normally used paraphyletically, outside of cladistics, e.g. as a 'grade', which are fruitless to precisely delineate, especially when including extinct species. Radiation results in the generation of new subclades by bifurcation, but in practice sexual hybridization may blur very closely related groupings.

In biology, phylogenetics is the study of the evolutionary history and relationships among or within groups of organisms. These relationships are determined by phylogenetic inference methods that focus on observed heritable traits, such as DNA sequences, protein amino acid sequences, or morphology. The result of such an analysis is a phylogenetic tree—a diagram containing a hypothesis of relationships that reflects the evolutionary history of a group of organisms.
In biology, phenetics, also known as taximetrics, is an attempt to classify organisms based on overall similarity, usually in morphology or other observable traits, regardless of their phylogeny or evolutionary relation. It is closely related to numerical taxonomy which is concerned with the use of numerical methods for taxonomic classification. Many people contributed to the development of phenetics, but the most influential were Peter Sneath and Robert R. Sokal. Their books are still primary references for this sub-discipline, although now out of print.

Biological systematics is the study of the diversification of living forms, both past and present, and the relationships among living things through time. Relationships are visualized as evolutionary trees. Phylogenies have two components: branching order and branch length. Phylogenetic trees of species and higher taxa are used to study the evolution of traits and the distribution of organisms (biogeography). Systematics, in other words, is used to understand the evolutionary history of life on Earth.

In biology, taxonomy is the scientific study of naming, defining (circumscribing) and classifying groups of biological organisms based on shared characteristics. Organisms are grouped into taxa and these groups are given a taxonomic rank; groups of a given rank can be aggregated to form a more inclusive group of higher rank, thus creating a taxonomic hierarchy. The principal ranks in modern use are domain, kingdom, phylum, class, order, family, genus, and species. The Swedish botanist Carl Linnaeus is regarded as the founder of the current system of taxonomy, as he developed a ranked system known as Linnaean taxonomy for categorizing organisms and binominal nomenclature for naming organisms.
Pattern recognition is the automated recognition of patterns and regularities in data. It has applications in statistical data analysis, signal processing, image analysis, information retrieval, bioinformatics, data compression, computer graphics and machine learning. Pattern recognition has its origins in statistics and engineering; some modern approaches to pattern recognition include the use of machine learning, due to the increased availability of big data and a new abundance of processing power. These activities can be viewed as two facets of the same field of application, and they have undergone substantial development over the past few decades.
Bloom's taxonomy is a set of three hierarchical models used for classification of educational learning objectives into levels of complexity and specificity. The three lists cover the learning objectives in cognitive, affective and psychomotor domains. The cognitive domain list has been the primary focus of most traditional education and is frequently used to structure curriculum learning objectives, assessments and activities.
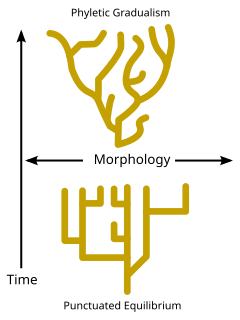
Phylogenesis is the biological process by which a taxon appears. The science that studies these processes is called phylogenetics.

Thomas (Tom) Cavalier-Smith, FRS, FRSC, NERC Professorial Fellow, was a Professor of Evolutionary Biology in the Department of Zoology, at the University of Oxford.

The history of plant systematics—the biological classification of plants—stretches from the work of ancient Greek to modern evolutionary biologists. As a field of science, plant systematics came into being only slowly, early plant lore usually being treated as part of the study of medicine. Later, classification and description was driven by natural history and natural theology. Until the advent of the theory of evolution, nearly all classification was based on the scala naturae. The professionalization of botany in the 18th and 19th century marked a shift toward more holistic classification methods, eventually based on evolutionary relationships.
Sentiment analysis is the use of natural language processing, text analysis, computational linguistics, and biometrics to systematically identify, extract, quantify, and study affective states and subjective information. Sentiment analysis is widely applied to voice of the customer materials such as reviews and survey responses, online and social media, and healthcare materials for applications that range from marketing to customer service to clinical medicine. With the rise of deep language models, such as RoBERTa, also more difficult data domains can be analyzed, e.g., news texts where authors typically express their opinion/sentiment less explicitly.
Robert Reuven Sokal was an Austrian-American biostatistician and entomologist. Distinguished Professor Emeritus at the Stony Brook University, Sokal was a member of the National Academy of Sciences and the American Academy of Arts and Sciences. He promoted the use of statistics in biology and co-founded the field of numerical taxonomy, together with Peter H. A. Sneath.
Walter Max Zimmermann was a German botanist and systematist. Zimmernann’s notions of classifying life objectively based on phylogenetic methods and on evolutionarily important characters were foundational for modern phylogenetics. Though they were later implemented by Willi Hennig in his fundamental work on phylogenetic systematics, Zimmermann's contributions to this field have largely been overlooked. Zimmermann also made several significant developments in the field of plant systematics such as the discovery of the telome theory. The standard botanical author abbreviation W.Zimm. is applied to species he described.
In biology, a phylum is a level of classification or taxonomic rank below kingdom and above class. Traditionally, in botany the term division has been used instead of phylum, although the International Code of Nomenclature for algae, fungi, and plants accepts the terms as equivalent. Depending on definitions, the animal kingdom Animalia contains about 31 phyla, the plant kingdom Plantae contains about 14 phyla, and the fungus kingdom Fungi contains about 8 phyla. Current research in phylogenetics is uncovering the relationships between phyla, which are contained in larger clades, like Ecdysozoa and Embryophyta.
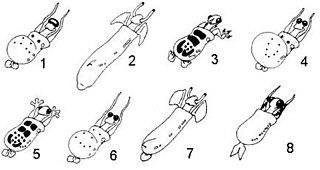
Caminalcules are a fictive group of animal-like life forms, which were created as a tool for better understanding phylogenetics in real organisms. They were created by Joseph H. Camin and consist of 29 living 'species' and 48 fossil forms.
An Operational Taxonomic Unit (OTU) is an operational definition used to classify groups of closely related individuals. The term was originally introduced in 1963 by Robert R. Sokal and Peter H. A. Sneath in the context of numerical taxonomy, where an "Operational Taxonomic Unit" is simply the group of organisms currently being studied. In this sense, an OTU is a pragmatic definition to group individuals by similarity, equivalent to but not necessarily in line with classical Linnaean taxonomy or modern evolutionary taxonomy.
Peter Henry Andrews Sneath FRS, MD was a microbiologist who co-founded the field of numerical taxonomy, together with Robert R. Sokal. Sneath and Sokal wrote Principles of Numerical Taxonomy, revised in 1973 as Numerical Taxonomy. Sneath reviewed the state of numerical taxonomy in 1995 and wrote some autobiographical notes in 2010.
Taxonomy is the practice and science of categorization or classification.
The term classification can apply to one or all of:
References
- ↑ "Numerical Taxonomy (biology)". www.accessscience.com. McGraw Hill Ltd. Retrieved 13 April 2010.
- ↑ Sokal & Sneath: Principles of Numerical Taxonomy, San Francisco: W.H. Freeman, 1963
- ↑ Sneath and Sokal: Numerical Taxonomy, San Francisco: W.H. Freeman, 1974 by Tejanshu Ravesh