
Gram-negative bacteria are bacteria that do not retain the crystal violet stain used in the Gram staining method of bacterial differentiation. They are characterized by their cell envelopes, which are composed of a thin peptidoglycan cell wall sandwiched between an inner cytoplasmic cell membrane and a bacterial outer membrane.

A transmembrane protein (TP) is a type of integral membrane protein that spans the entirety of the cell membrane. Many transmembrane proteins function as gateways to permit the transport of specific substances across the membrane. They frequently undergo significant conformational changes to move a substance through the membrane. They are usually highly hydrophobic and aggregate and precipitate in water. They require detergents or nonpolar solvents for extraction, although some of them (beta-barrels) can be also extracted using denaturing agents.
Braun's lipoprotein, found in some gram-negative cell walls, is one of the most abundant membrane proteins; its molecular weight is about 7.2 kDa. It is bound at its C-terminal end by a covalent bond to the peptidoglycan layer and is embedded in the outer membrane by its hydrophobic head. BLP tightly links the two layers and provides structural integrity to the outer membrane.

Porins are beta barrel proteins that cross a cellular membrane and act as a pore, through which molecules can diffuse. Unlike other membrane transport proteins, porins are large enough to allow passive diffusion, i.e., they act as channels that are specific to different types of molecules. They are present in the outer membrane of gram-negative bacteria and some gram-positive mycobacteria, the outer membrane of mitochondria, and the outer chloroplast membrane.

The bacterial outer membrane is found in gram-negative bacteria. Its composition is distinct from that of the inner cytoplasmic cell membrane - among other things, the outer leaflet of the outer membrane of many gram-negative bacteria includes a complex lipopolysaccharide whose lipid portion acts as an endotoxin - and in some bacteria such as E. coli it is linked to the cell's peptidoglycan by Braun's lipoprotein.
Bacterial display is a protein engineering technique used for in vitro protein evolution. Libraries of polypeptides displayed on the surface of bacteria can be screened using flow cytometry or iterative selection procedures (biopanning). This protein engineering technique allows us to link the function of a protein with the gene that encodes it. Bacterial display can be used to find target proteins with desired properties and can be used to make affinity ligands which are cell-specific. This system can be used in many applications including the creation of novel vaccines, the identification of enzyme substrates and finding the affinity of a ligand for its target protein.
The bacterium, despite its simplicity, contains a well-developed cell structure which is responsible for some of its unique biological structures and pathogenicity. Many structural features are unique to bacteria and are not found among archaea or eukaryotes. Because of the simplicity of bacteria relative to larger organisms and the ease with which they can be manipulated experimentally, the cell structure of bacteria has been well studied, revealing many biochemical principles that have been subsequently applied to other organisms.

A colicin is a type of bacteriocin produced by and toxic to some strains of Escherichia coli. Colicins are released into the environment to reduce competition from other bacterial strains. Colicins bind to outer membrane receptors, using them to translocate to the cytoplasm or cytoplasmic membrane, where they exert their cytotoxic effect, including depolarisation of the cytoplasmic membrane, DNase activity, RNase activity, or inhibition of murein synthesis.

Voltage-dependent anion channels, or mitochondrial porins, are a class of porin ion channel located on the outer mitochondrial membrane. There is debate as to whether or not this channel is expressed in the cell surface membrane.

General bacterial porins are a family of proteins from the outer membranes of Gram-negative bacteria. The porins act as molecular filters for hydrophilic compounds. They are responsible for the 'molecular sieve' properties of the outer membrane. Porins form large water-filled channels which allow the diffusion of hydrophilic molecules into the periplasmic space. Some porins form general diffusion channels that allow any solute up to a certain size to cross the membrane, while other porins are specific for one particular solute and contain a binding site for that solute inside the pores. As porins are the major outer membrane proteins, they also serve as receptor sites for the binding of phages and bacteriocins.

Outer membrane receptors, also known as TonB-dependent receptors, are a family of beta barrel proteins named for their localization in the outer membrane of gram-negative bacteria. TonB complexes sense signals from the outside of bacterial cells and transmit them into the cytoplasm, leading to transcriptional activation of target genes. TonB-dependent receptors in gram-negative bacteria are associated with the uptake and transport of large substrates such as iron siderophore complexes and vitamin B12.
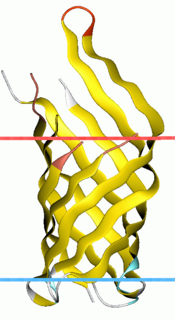
OmpA-like transmembrane domain is an evolutionarily conserved domain of bacterial outer membrane proteins. This domain consists of an eight-stranded beta barrel. OmpA is the predominant cell surface antigen in enterobacteria found in about 100,000 copies per cell. The expression of OmpA is tightly regulated by a variety of mechanisms. One mechanism by which OmpA expression is regulated in Vibrio species is by an antisense non-coding RNA called VrrA.
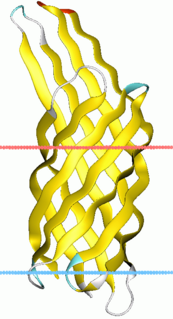
Virulence-related outer membrane proteins are expressed in the outer membrane of gram-negative bacteria and are essential to bacterial survival within macrophages and for eukaryotic cell invasion.

Outer membrane protein G (OmpG) is a porin, a channel proteins in the outer membrane of Gram-negative bacteria.
Motility protein A, also known as MotA Pait, is a bacterial protein that is encoded by the motA gene. It is a component of the flagellar motor. More specifically, MotA and MotB make the stator of a H+ driven bacterial flagellum and surround the rotor as a ring of about 8–10 particles. MotA and MotB are integral membrane proteins. MotA has four transmembrane domains.
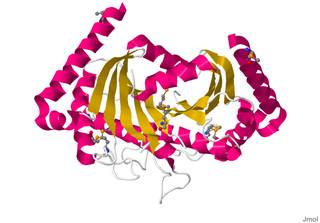
Motility protein B also known as MotB is a bacterial protein that is encoded by the motB gene. It's a component of the flagellar motor. More specifically, MotA and MotB makes the stator of a flagellum and surround the rotor as a ring of about 8-10 particles. MotA and MotB are integral membrane proteins. While both MotA and MotB surround the MS ring, MotB also anchors MotA to cell wall peptidoglycan. These two proteins form pores that harvest energy for flagellar mechanical movement by proton motive force (PMF) across the membrane. Cellular metabolic processes such as the electron transport chain move protons outside the cell, creating more protons and more positive charge in the extracellular space. When the protons flow back into the cell through MotA and MotB along concentration and charge gradients, they release energy that is used for flagellar rotation. The speed of the flagellar motor is dependent on the magnitude of the PMF acting on MotA and MotB.
Omptins are a family of bacterial proteases. They are aspartate proteases, which cleave peptides with the use of a water molecule. Found in the outer membrane of gram-negative enterobacteria such as Shigella flexneri, Yersinia pestis, Escherichia coli, and Salmonella enterica. Omptins consist of a widely conserved beta barrel spanning the membrane with 5 extracellular loops. These loops are responsible for the various substrate specificities. These proteases rely upon binding of lipopolysaccharide for activity.
EnvZ/OmpR is a two-component regulatory system widely distributed in bacteria and particularly well characterized in Escherichia coli. Its function is in osmoregulation, responding to changes in environmental osmolality by regulating the expression of the outer membrane porins OmpF and OmpC. EnvZ is a histidine kinase which also possesses a cytoplasmic osmosensory domain, and OmpR is its corresponding response regulator protein.

In molecular biology, YadA is a protein domain which is short for Yersinia adhesin A. These proteins have strong sequence and structural homology, particularly at their C-terminal end. The function is to promote their pathogenicity and virulence in host cells, though cell adhesion. YadA is found in three pathogenic species of Yersinia, Y. pestis,Y. pseudotuberculosis, and Y. enterocolitica. The YadA domain is encoded for by a virulence plasmid in Yersinia, which encodes a type-III secretion (T3S) system consisting of the Ysc injectisome and the Yop effectors.

OmpT is an aspartyl protease found on the outer membrane of Escherichia coli. OmpT is a subtype of the family of omptin proteases, which are found on some gram-negative species of bacteria.














