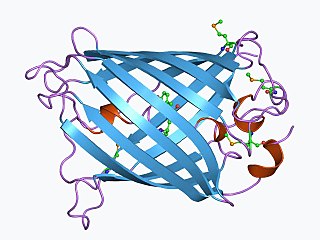
The green fluorescent protein (GFP) is a protein that exhibits bright green fluorescence when exposed to light in the blue to ultraviolet range. The label GFP traditionally refers to the protein first isolated from the jellyfish Aequorea victoria and is sometimes called avGFP. However, GFPs have been found in other organisms including corals, sea anemones, zoanithids, copepods and lancelets.

Bioluminescence is the production and emission of light by living organisms. It is a form of chemiluminescence. Bioluminescence occurs widely in marine vertebrates and invertebrates, as well as in some fungi, microorganisms including some bioluminescent bacteria, and terrestrial arthropods such as fireflies. In some animals, the light is bacteriogenic, produced by symbiotic bacteria such as those from the genus Vibrio; in others, it is autogenic, produced by the animals themselves.

In molecular biology and biotechnology, a fluorescent tag, also known as a fluorescent label or fluorescent probe, is a molecule that is attached chemically to aid in the detection of a biomolecule such as a protein, antibody, or amino acid. Generally, fluorescent tagging, or labeling, uses a reactive derivative of a fluorescent molecule known as a fluorophore. The fluorophore selectively binds to a specific region or functional group on the target molecule and can be attached chemically or biologically. Various labeling techniques such as enzymatic labeling, protein labeling, and genetic labeling are widely utilized. Ethidium bromide, fluorescein and green fluorescent protein are common tags. The most commonly labelled molecules are antibodies, proteins, amino acids and peptides which are then used as specific probes for detection of a particular target.
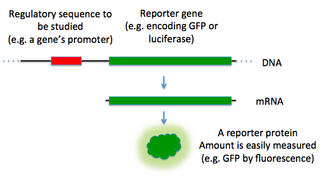
In molecular biology, a reporter gene is a gene that researchers attach to a regulatory sequence of another gene of interest in bacteria, cell culture, animals or plants. Such genes are called reporters because the characteristics they confer on organisms expressing them are easily identified and measured, or because they are selectable markers. Reporter genes are often used as an indication of whether a certain gene has been taken up by or expressed in the cell or organism population.
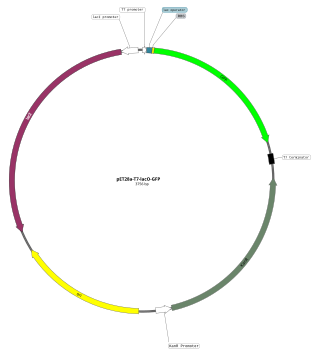
An expression vector, otherwise known as an expression construct, is usually a plasmid or virus designed for gene expression in cells. The vector is used to introduce a specific gene into a target cell, and can commandeer the cell's mechanism for protein synthesis to produce the protein encoded by the gene. Expression vectors are the basic tools in biotechnology for the production of proteins.

Aequorea victoria, also sometimes called the crystal jelly, is a bioluminescent hydrozoan jellyfish, or hydromedusa, that is found off the west coast of North America.

Alba was the name of a genetically modified "glowing" rabbit created as an artistic work by contemporary artist Eduardo Kac, produced in collaboration with French geneticist Louis-Marie Houdebine.
In biology, a marker gene may have several meanings. In nuclear biology and molecular biology, a marker gene is a gene used to determine if a nucleic acid sequence has been successfully inserted into an organism's DNA. In particular, there are two sub-types of these marker genes: a selectable marker and a marker for screening. In metagenomics and phylogenetics, a marker gene is an orthologous gene group which can be used to delineate between taxonomic lineages.

Aequorin is a calcium-activated photoprotein isolated from the hydrozoan Aequorea victoria. Its bioluminescence was studied decades before the protein was isolated from the animal by Osamu Shimomura in 1962. In the animal, the protein occurs together with the green fluorescent protein to produce green light by resonant energy transfer, while aequorin by itself generates blue light.
The L-arabinose operon, also called the ara or araBAD operon, is an operon required for the breakdown of the five-carbon sugar L-arabinose in Escherichia coli. The L-arabinose operon contains three structural genes: araB, araA, araD, which encode for three metabolic enzymes that are required for the metabolism of L-arabinose. AraB (ribulokinase), AraA, and AraD produced by these genes catalyse conversion of L-arabinose to an intermediate of the pentose phosphate pathway, D-xylulose-5-phosphate.

Roger Yonchien Tsien was an American biochemist. He was a professor of chemistry and biochemistry at the University of California, San Diego and was awarded the Nobel Prize in Chemistry in 2008 for his discovery and development of the green fluorescent protein, in collaboration with organic chemist Osamu Shimomura and neurobiologist Martin Chalfie. Tsien was also a pioneer of calcium imaging.

Aequorea forskalea is a species of hydrozoan in the family Aequoreidae. Discovered in 1810 by Péron and Lesueur, A. forskalea was initially found in coastal to offshore waters of the Mediterranean Sea. This species is commonly referred to as the many-ribbed jellyfish. The species is often mixed up with some other members of the genus due to some similarities including the capability of bioluminescence.
In molecular cloning, a vector is any particle used as a vehicle to artificially carry a foreign nucleic sequence – usually DNA – into another cell, where it can be replicated and/or expressed. A vector containing foreign DNA is termed recombinant DNA. The four major types of vectors are plasmids, viral vectors, cosmids, and artificial chromosomes. Of these, the most commonly used vectors are plasmids. Common to all engineered vectors are an origin of replication, a multicloning site, and a selectable marker.

Bioreporters are intact, living microbial cells that have been genetically engineered to produce a measurable signal in response to a specific chemical or physical agent in their environment. Bioreporters contain two essential genetic elements, a promoter gene and a reporter gene. The promoter gene is turned on (transcribed) when the target agent is present in the cell’s environment. The promoter gene in a normal bacterial cell is linked to other genes that are then likewise transcribed and then translated into proteins that help the cell in either combating or adapting to the agent to which it has been exposed. In the case of a bioreporter, these genes, or portions thereof, have been removed and replaced with a reporter gene. Consequently, turning on the promoter gene now causes the reporter gene to be turned on. Activation of the reporter gene leads to production of reporter proteins that ultimately generate some type of a detectable signal. Therefore, the presence of a signal indicates that the bioreporter has sensed a particular target agent in its environment.

Osamu Shimomura was a Japanese organic chemist and marine biologist, and professor emeritus at Marine Biological Laboratory (MBL) in Woods Hole, Massachusetts and Boston University School of Medicine. He was awarded the Nobel Prize in Chemistry in 2008 for the discovery and development of green fluorescent protein (GFP) with two American scientists: Martin Chalfie of Columbia University and Roger Tsien of the University of California-San Diego.

Martin Lee Chalfie is an American scientist. He is University Professor at Columbia University. He shared the 2008 Nobel Prize in Chemistry along with Osamu Shimomura and Roger Y. Tsien "for the discovery and development of the green fluorescent protein, GFP". He holds a PhD in neurobiology from Harvard University.
Douglas C. Prasher is an American molecular biologist. He is known for his work to clone and sequence the genes for the photoprotein aequorin and green fluorescent protein (GFP) and for his proposal to use GFP as a tracer molecule. He communicated his pioneering work to Martin Chalfie and Roger Y. Tsien, but by 1991 he was unable to obtain further research funding, and left academia. Eventually, he had to abandon science. Chalfie and Tsien were awarded the 2008 Nobel Prize in Chemistry for work that they publicly acknowledged was substantially based on Prasher's work; through their efforts and those of others, he returned to scientific research in June 2010.

Coelenteramide is the oxidized product, or oxyluciferin, of the bioluminescent reactions in many marine organisms that use coelenterazine. It was first isolated as a blue fluorescent protein from Aequorea victoria after the animals were stimulated to emit light. Under basic conditions, the compound will break down further into coelenteramine and 4-hydroxyphenylacetic acid.

John Woodland "Woody" Hastings, was a leader in the field of photobiology, especially bioluminescence, and was one of the founders of the field of circadian biology. He was the Paul C. Mangelsdorf Professor of Natural Sciences and Professor of Molecular and Cellular Biology at Harvard University. He published over 400 papers and co-edited three books.
mCherry is a member of the mFruits family of monomeric red fluorescent proteins (mRFPs). As a RFP, mCherry was derived from DsRed of Discosoma sea anemones unlike green fluorescent proteins (GFPs) which are often derived from Aequorea victoria jellyfish. Fluorescent proteins are used to tag components in the cell, so they can be studied using fluorescence spectroscopy and fluorescence microscopy. mCherry absorbs light between 540-590 nm and emits light in the range of 550-650 nm. mCherry belongs to the group of fluorescent protein chromophores used as instruments to visualize genes and analyze their functions in experiments. Genome editing has been improved greatly through the precise insertion of these fluorescent protein tags into the genetic material of many diverse organisms. Most comparisons between the brightness and photostability of different fluorescent proteins have been made in vitro, removed from biological variables that affect protein performance in cells or organisms. It is hard to perfectly simulate cellular environments in vitro, and the difference in environment could have an effect on the brightness and photostability.