
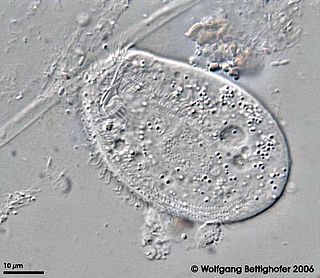
Paramecium is a genus of eukaryotic, unicellular ciliates, commonly studied as a representative of the ciliate group. Paramecia are widespread in freshwater, brackish, and marine environments and are often very abundant in stagnant basins and ponds. Because some species are readily cultivated and easily induced to conjugate and divide, it has been widely used in classrooms and laboratories to study biological processes. Its usefulness as a model organism has caused one ciliate researcher to characterize it as the "white rat" of the phylum Ciliophora.
In genetics, an expressed sequence tag (EST) is a short sub-sequence of a cDNA sequence. ESTs may be used to identify gene transcripts, and were instrumental in gene discovery and in gene-sequence determination. The identification of ESTs has proceeded rapidly, with approximately 74.2 million ESTs now available in public databases. EST approaches have largely been superseded by whole genome and transcriptome sequencing and metagenome sequencing.

In genetics, a single-nucleotide polymorphism is a germline substitution of a single nucleotide at a specific position in the genome. Although certain definitions require the substitution to be present in a sufficiently large fraction of the population, many publications do not apply such a frequency threshold.

Sequence homology is the biological homology between DNA, RNA, or protein sequences, defined in terms of shared ancestry in the evolutionary history of life. Two segments of DNA can have shared ancestry because of three phenomena: either a speciation event (orthologs), or a duplication event (paralogs), or else a horizontal gene transfer event (xenologs).
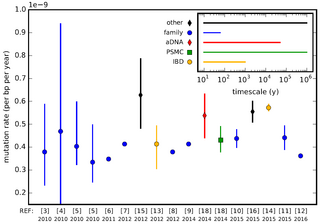
In genetics, the mutation rate is the frequency of new mutations in a single gene or organism over time. Mutation rates are not constant and are not limited to a single type of mutation, therefore there are many different types of mutations. Mutation rates are given for specific classes of mutations. Point mutations are a class of mutations which are changes to a single base. Missense and Nonsense mutations are two subtypes of point mutations. The rate of these types of substitutions can be further subdivided into a mutation spectrum which describes the influence of the genetic context on the mutation rate.
Tracy Morton Sonneborn was an American biologist. His life's study was of the protozoan group Paramecium.

The synaptonemal complex (SC) is a protein structure that forms between homologous chromosomes during meiosis and is thought to mediate synapsis and recombination during meiosis I in eukaryotes. It is currently thought that the SC functions primarily as a scaffold to allow interacting chromatids to complete their crossover activities.
Microbial genetics is a subject area within microbiology and genetic engineering. Microbial genetics studies microorganisms for different purposes. The microorganisms that are observed are bacteria, and archaea. Some fungi and protozoa are also subjects used to study in this field. The studies of microorganisms involve studies of genotype and expression system. Genotypes are the inherited compositions of an organism. Genetic Engineering is a field of work and study within microbial genetics. The usage of recombinant DNA technology is a process of this work. The process involves creating recombinant DNA molecules through manipulating a DNA sequence. That DNA created is then in contact with a host organism. Cloning is also an example of genetic engineering.

The Generic Model Organism Database (GMOD) project provides biological research communities with a toolkit of open-source software components for visualizing, annotating, managing, and storing biological data. The GMOD project is funded by the United States National Institutes of Health, National Science Foundation and the USDA Agricultural Research Service.

RNA-dependent RNA polymerase (RdRp) or RNA replicase is an enzyme that catalyzes the replication of RNA from an RNA template. Specifically, it catalyzes synthesis of the RNA strand complementary to a given RNA template. This is in contrast to typical DNA-dependent RNA polymerases, which all organisms use to catalyze the transcription of RNA from a DNA template.
Phycodnaviridae is a family of large (100–560 kb) double-stranded DNA viruses that infect marine or freshwater eukaryotic algae. Viruses within this family have a similar morphology, with an icosahedral capsid. As of 2014, there were 33 species in this family, divided among 6 genera. This family belongs to a super-group of large viruses known as nucleocytoplasmic large DNA viruses. Evidence was published in 2014 suggesting that specific strains of Phycodnaviridae might infect humans rather than just algal species, as was previously believed. Most genera under this family enter the host cell by cell receptor endocytosis and replicate in the nucleus. Phycodnaviridae play important ecological roles by regulating the growth and productivity of their algal hosts. Algal species such Heterosigma akashiwo and the genus Chrysochromulina can form dense blooms which can be damaging to fisheries, resulting in losses in the aquaculture industry. Heterosigma akashiwo virus (HaV) has been suggested for use as a microbial agent to prevent the recurrence of toxic red tides produced by this algal species. Phycodnaviridae cause death and lysis of freshwater and marine algal species, liberating organic carbon, nitrogen and phosphorus into the water, providing nutrients for the microbial loop.

The Single Nucleotide Polymorphism Database (dbSNP) is a free public archive for genetic variation within and across different species developed and hosted by the National Center for Biotechnology Information (NCBI) in collaboration with the National Human Genome Research Institute (NHGRI). Although the name of the database implies a collection of one class of polymorphisms only, it in fact contains a range of molecular variation: (1) SNPs, (2) short deletion and insertion polymorphisms (indels/DIPs), (3) microsatellite markers or short tandem repeats (STRs), (4) multinucleotide polymorphisms (MNPs), (5) heterozygous sequences, and (6) named variants. The dbSNP accepts apparently neutral polymorphisms, polymorphisms corresponding to known phenotypes, and regions of no variation. It was created in September 1998 to supplement GenBank, NCBI’s collection of publicly available nucleic acid and protein sequences.
The UCSC Genome Browser is an online and downloadable genome browser hosted by the University of California, Santa Cruz (UCSC). It is an interactive website offering access to genome sequence data from a variety of vertebrate and invertebrate species and major model organisms, integrated with a large collection of aligned annotations. The Browser is a graphical viewer optimized to support fast interactive performance and is an open-source, web-based tool suite built on top of a MySQL database for rapid visualization, examination, and querying of the data at many levels. The Genome Browser Database, browsing tools, downloadable data files, and documentation can all be found on the UCSC Genome Bioinformatics website.

Paramecium aurelia are unicellular organisms belonging to the genus Paramecium of the phylum Ciliophora. They are covered in cilia which help in movement and feeding.Paramecium can reproduce sexually, asexually, or by the process of endomixis. Paramecium aurelia demonstrate a strong “sex reaction” whereby groups of individuals will cluster together, and emerge in conjugant pairs. This pairing can last up to 12 hours, during which the micronucleus of each organism will be exchanged. In Paramecium aurelia, a cryptic species complex was discovered by observation. Since then, some have tried to decode this complex using genetic data.
Chilodonella uncinata is a single-celled organism of the ciliate class of alveoles. As a ciliate, C. uncinata has cilia covering its body and a dual nuclear structure, the micronucleus and macronucleus. Unlike some other ciliates, C. uncinata contains millions of minichromosomes in its macronucleus while its micronucleus is estimated to contain 3 chromosomes. Childonella uncinata is the causative agent of Chilodonelloza, a disease that affects the gills and skin of fresh water fish, and may act as a facultative parasite of mosquito larva.

Sterkiella histriomuscorum, formerly Oxytricha trifallax, is a ciliate species in the genus Sterkiella, known for its highly fragmented genomes which have been used as a model for ciliate genetics.
The mold, protozoan, and coelenterate mitochondrial code and the mycoplasma/spiroplasma code is the genetic code used by various organisms, in some cases with slight variations, notably the use of UGA as a tryptophan codon rather than a stop codon.
Autogamy, or self-fertilization, refers to the fusion of two gametes that come from one individual. Autogamy is predominantly observed in the form of self-pollination, a reproductive mechanism employed by many flowering plants. However, species of protists have also been observed using autogamy as a means of reproduction. Flowering plants engage in autogamy regularly, while the protists that engage in autogamy only do so in stressful environments.
Gene amplification in Paramecium tetraurelia is an example of gene amplification that has occurred in the unicellular organism Paramecium tetraurelia.









