
In immunology, an antigen (Ag) is any molecule, molecular structure, foreign particulate matter, or pollen grain that can bind to a specific antibody or T-cell receptor. The presence of antigens in the body may trigger an immune response. Antigens can be proteins, peptides, polysaccharides, lipids, or nucleic acids.
Combinatorial chemistry comprises chemical synthetic methods that make it possible to prepare a large number of compounds in a single process. These compound libraries can be made as mixtures, sets of individual compounds or chemical structures generated by computer software. Combinatorial chemistry can be used for the synthesis of small molecules and for peptides.
An epitope, also known as antigenic determinant, is the part of an antigen that is recognized by the immune system, specifically by antibodies, B cells, or T cells. The part of an antibody that binds to the epitope is called a paratope. Although epitopes are usually non-self proteins, sequences derived from the host that can be recognized are also epitopes.
A cancer vaccine is a vaccine that either treats existing cancer or prevents development of cancer. Vaccines that treat existing cancer are known as therapeutic cancer vaccines or tumor antigen vaccines. Some of the vaccines are "autologous", being prepared from samples taken from the patient, and are specific to that patient.
Polyclonal antibodies (pAbs) are antibodies that are secreted by different B cell lineages within the body. They are a collection of immunoglobulin molecules that react against a specific antigen, each identifying a different epitope.
Antigen processing, or the cytosolic pathway, is an immunological process that prepares antigens for presentation to special cells of the immune system called T lymphocytes. It is considered to be a stage of antigen presentation pathways. This process involves two distinct pathways for processing of antigens from an organism's own (self) proteins or intracellular pathogens, or from phagocytosed pathogens ; subsequent presentation of these antigens on class I or class II major histocompatibility complex (MHC) molecules is dependent on which pathway is used. Both MHC class I and II are required to bind antigen before they are stably expressed on a cell surface. MHC I antigen presentation typically involves the endogenous pathway of antigen processing, and MHC II antigen presentation involves the exogenous pathway of antigen processing. Cross-presentation involves parts of the exogenous and the endogenous pathways but ultimately involves the latter portion of the endogenous pathway.

In immunology, epitope mapping is the process of experimentally identifying the binding site, or epitope, of an antibody on its target antigen. Identification and characterization of antibody binding sites aid in the discovery and development of new therapeutics, vaccines, and diagnostics. Epitope characterization can also help elucidate the binding mechanism of an antibody and can strengthen intellectual property (patent) protection. Experimental epitope mapping data can be incorporated into robust algorithms to facilitate in silico prediction of B-cell epitopes based on sequence and/or structural data.
Bacterial display is a protein engineering technique used for in vitro protein evolution. Libraries of polypeptides displayed on the surface of bacteria can be screened using flow cytometry or iterative selection procedures (biopanning). This protein engineering technique allows us to link the function of a protein with the gene that encodes it. Bacterial display can be used to find target proteins with desired properties and can be used to make affinity ligands which are cell-specific. This system can be used in many applications including the creation of novel vaccines, the identification of enzyme substrates and finding the affinity of a ligand for its target protein.
FLAG-tag, or FLAG octapeptide, or FLAG epitope, is a peptide protein tag that can be added to a protein using recombinant DNA technology, having the sequence DYKDDDDK. It is one of the most specific tags and it is an artificial antigen to which specific, high affinity monoclonal antibodies have been developed and hence can be used for protein purification by affinity chromatography and also can be used for locating proteins within living cells. FLAG-tag has been used to separate recombinant, overexpressed protein from wild-type protein expressed by the host organism. FLAG-tag can also be used in the isolation of protein complexes with multiple subunits, because FLAG-tag's mild purification procedure tends not to disrupt such complexes. FLAG-tag-based purification has been used to obtain proteins of sufficient purity and quality to carry out 3D structure determination by x-ray crystallography.
A peptide library is a tool for studying proteins. Peptide libraries contain a great number of peptides that have a systematic combination of amino acids. Usually, the peptide library is synthesized on a solid phase, mostly on resin, which can be made as a flat surface or beads. The peptide library provides a powerful tool for drug design, protein–protein interactions, and other biochemical and pharmaceutical applications.
A mimotope is often a peptide, and mimics the structure of an epitope. Because of this property it causes an antibody response similar to the one elicited by the epitope. An antibody for a given epitope antigen will recognize a mimotope which mimics that epitope. Mimotopes are commonly obtained from phage display libraries through biopanning. Vaccines utilizing mimotopes are being developed. Mimotopes are a kind of peptide aptamers.
Computational Resources for Drug Discovery (CRDD) is one of the important silico modules of Open Source for Drug Discovery (OSDD). The CRDD web portal provides computer resources related to drug discovery on a single platform. It provides computational resources for researchers in computer-aided drug design, a discussion forum, and resources to maintain Wikipedia related to drug discovery, predict inhibitors, and predict the ADME-Tox property of molecules One of the major objectives of CRDD is to promote open source software in the field of chemoinformatics and pharmacoinformatics.
Peptide-based synthetic vaccines, also called epitope vaccines, are subunit vaccines made from peptides. The peptides mimic the epitopes of the antigen that triggers direct or potent immune responses. Peptide vaccines can not only induce protection against infectious pathogens and non-infectious diseases but also be utilized as therapeutic cancer vaccines, where peptides from tumor-associated antigens are used to induce an effective anti-tumor T-cell response.
Immunomics is the study of immune system regulation and response to pathogens using genome-wide approaches. With the rise of genomic and proteomic technologies, scientists have been able to visualize biological networks and infer interrelationships between genes and/or proteins; recently, these technologies have been used to help better understand how the immune system functions and how it is regulated. Two thirds of the genome is active in one or more immune cell types and less than 1% of genes are uniquely expressed in a given type of cell. Therefore, it is critical that the expression patterns of these immune cell types be deciphered in the context of a network, and not as an individual, so that their roles be correctly characterized and related to one another. Defects of the immune system such as autoimmune diseases, immunodeficiency, and malignancies can benefit from genomic insights on pathological processes. For example, analyzing the systematic variation of gene expression can relate these patterns with specific diseases and gene networks important for immune functions.
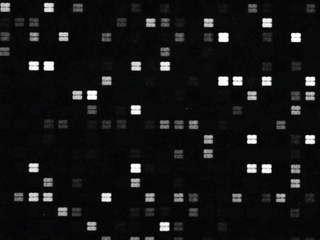
A peptide microarray is a collection of peptides displayed on a solid surface, usually a glass or plastic chip. Peptide chips are used by scientists in biology, medicine and pharmacology to study binding properties and functionality and kinetics of protein-protein interactions in general. In basic research, peptide microarrays are often used to profile an enzyme, to map an antibody epitope or to find key residues for protein binding. Practical applications are seromarker discovery, profiling of changing humoral immune responses of individual patients during disease progression, monitoring of therapeutic interventions, patient stratification and development of diagnostic tools and vaccines.
Immunodominance is the immunological phenomenon in which immune responses are mounted against only a few of the antigenic peptides out of the many produced. That is, despite multiple allelic variations of MHC molecules and multiple peptides presented on antigen presenting cells, the immune response is skewed to only specific combinations of the two. Immunodominance is evident for both antibody-mediated immunity and cell-mediated immunity. Epitopes that are not targeted or targeted to a lower degree during an immune response are known as subdominant epitopes. The impact of immunodominance is immunodomination, where immunodominant epitopes will curtail immune responses against non-dominant epitopes. Antigen-presenting cells such as dendritic cells, can have up to six different types of MHC molecules for antigen presentation. There is a potential for generation of hundreds to thousands of different peptides from the proteins of pathogens. Yet, the effector cell population that is reactive against the pathogen is dominated by cells that recognize only a certain class of MHC bound to only certain pathogen-derived peptides presented by that MHC class. Antigens from a particular pathogen can be of variable immunogenicity, with the antigen that stimulates the strongest response being the immunodominant one. The different levels of immunogenicity amongst antigens forms what is known as dominance hierarchy.

The Rapid Deployment Vaccine Collaborative (RaDVaC), is a non-profit, collaborative, open-source vaccine research organization founded in March 2020 by Preston Estep and colleagues from various fields of expertise, motivated to respond to the COVID-19 pandemic through rapid, adaptable, transparent, and accessible vaccine development. The members of RaDVaC contend that even the accelerated vaccine approvals, such as the FDA's Emergency Use Authorization, does not make vaccines available quickly enough. The core group has published a series of white papers online, detailing both the technical principles of and protocols for their research vaccine formulas, as well as dedicated materials and protocols pages. All of the organization's published work has been released under Creative Commons non-commercial licenses, including those contributing to the Open COVID Pledge. Multiple individuals involved with the project have engaged in self-experimentation to assess vaccine safety and efficacy. As of January 2022, the organization has developed and published twelve iterations of experimental intranasal, multivalent, multi-epitope peptide vaccine formulas, and according to the RaDVaC website, by early 2021 hundreds of individuals had self-administered one or more doses of the vaccines described by the group.

Günther Jung is a German chemist. He was professor for Organic Chemistry and Biochemistry at the University of Tübingen from 1973 to 2002.

EpiVacCorona is a peptide-based vaccine against COVID-19 developed by the VECTOR center of Virology. The lack of protective effectiveness of EpiVacCorona, which is still in use in Russia, has been reported in scientific literature and in the media. The vaccine consists of three chemically synthesized peptides that are conjugated to a large carrier protein. This protein is a fusion product of a viral nucleocapsid protein and a bacterial MBP protein. A phase three clinical trial to show whether or not the vaccine can protect people against COVID-19 was launched in November 2020 with more than three thousand participants. The facts of the end of the trial and the result of the trial have not been made public.

UB-612 is a COVID-19 vaccine candidate developed by United Biomedical, Inc. Asia, and Vaxxinity, Inc. It is a peptide vaccine.






