
Flaviviridae is a family of viruses. Humans and other mammals serve as natural hosts. They are primarily spread through arthropod vectors. The family gets its name from the yellow fever virus, the type virus of Flaviviridae; flavus is Latin for "yellow", and yellow fever in turn was named because of its propensity to cause jaundice in victims. Currently, over 100 species are in this family, divided among four genera. Diseases associated with this family include: hepatitis (hepaciviruses), hemorrhagic syndromes, fatal mucosal disease (pestiviruses), hemorrhagic fever, encephalitis, and the birth defect microcephaly (flaviviruses).
The 5′ untranslated region is the region of an mRNA that is directly upstream from the initiation codon. This region is important for the regulation of translation of a transcript by differing mechanisms in viruses, prokaryotes and eukaryotes. While called untranslated, the 5′ UTR or a portion of it is sometimes translated into a protein product. This product can then regulate the translation of the main coding sequence of the mRNA. In many organisms, however, the 5′ UTR is completely untranslated, instead forming complex secondary structure to regulate translation. The 5′ UTR has been found to interact with proteins relating to metabolism; and proteins translate sequences within the 5′ UTR. In addition, this region has been involved in transcription regulation, such as the sex-lethal gene in Drosophila. Regulatory elements within 5′ UTRs have also been linked to mRNA export.
Dicistroviridae is a family of viruses in the order Picornavirales. Invertebrates, including aphids, leafhoppers, flies, bees, ants, and silkworms, serve as natural hosts. There are currently 15 species in this family, divided among 2 genera. Diseases associated with this family include: DCV: increased reproductive potential. extremely pathogenic when injected with high associated mortality. CrPV: paralysis and death.
Eukaryotic translation is the biological process by which messenger RNA is translated into proteins in eukaryotes. It consists of four phases: initiation, elongation, termination, and recycling.

This family represents the internal ribosome entry site (IRES) of the Picornaviruses. IRES elements allow cap and end-independent translation of mRNA in the host cell. The IRES achieves this by mediating the internal initiation of translation by recruiting a ribosomal 43S pre-initiation complex directly to the initiation codon and eliminates the requirement for the eukaryotic initiation factor eIF4F.

The BiP internal ribosome entry site (IRES) is an RNA element present in the 5' UTR of the mRNA of BiP protein and allows cap-independent translation. BiP protein expression has been found to be significantly enhanced by the heat shock response due to internal ribosome entry site (IRES)-dependent translation. It is thought that this translational mechanism is essential for the survival of cells under stress.

The Cripavirus internal ribosome entry site is an RNA element required for the production of capsid proteins through ribosome recruitment to an intergenic region IRES.
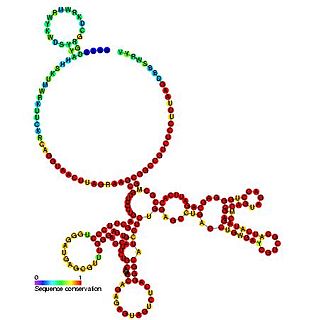
The Epstein–Barr virus nuclear-antigen internal ribosome entry site is an internal ribosome entry site (IRES) that is found in an exon in the 5' untranslated region of the Epstein–Barr virus nuclear antigen 1 (EBNA1) gene. The EBNA IRES allows EBNA1 translation to occur under situations where initiation from the 5' cap structure and ribosome scanning is reduced. It is thought that the EBNA IRES is necessary for the regulation of latent-gene expression.

This family represents the internal ribosome entry site (IRES) of the hepatitis A virus. HAV IRES is a 450 nucleotide long sequence located in the 735 nt long 5’ UTR of Hepatitis A viral RNA genome. IRES elements allow cap and end-independent translation of mRNA in the host cell. The IRES achieves this by mediating the internal initiation of translation by recruiting a ribosomal 40S pre-initiation complex directly to the initiation codon and eliminates the requirement for eukaryotic initiation factor, eIF4F.
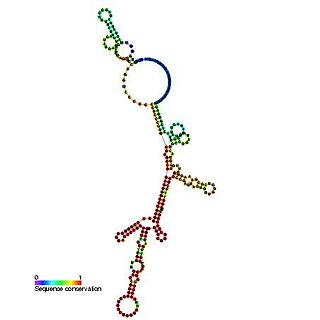
The Hepatitis C virus internal ribosome entry site, or HCV IRES, is an RNA structure within the 5'UTR of the HCV genome that mediates cap-independent translation initiation.
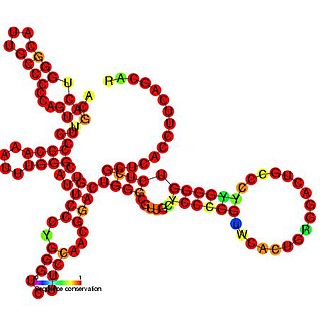
The insulin-like growth factor II (IGF-II) internal ribosome entry site IRES is found in the 5' UTR of IGF-II leader 2 mRNA. This RNA element allows cap-independent translation of the mRNA and it is thought that this family may facilitate a continuous IGF-II production in rapidly dividing cells during development. Ribosomal scanning on human insulin-like growth factor II (IGF-II) is hard to comprehend due to one open reading frame and the ability for the hormone to fold into a stable structure.
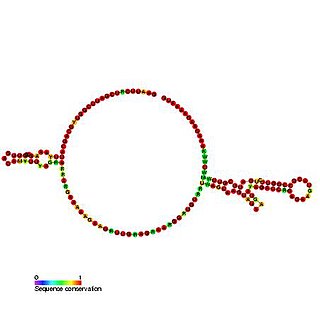
The Tobamovirus internal ribosome entry site (IRES) is an element that allows cap and end-independent translation of mRNA in the host cell. The IRES achieves this by mediating the internal initiation of translation by recruiting a ribosomal 43S pre-initiation complex directly to the initiation codon and eliminates the requirement for the eukaryotic initiation factor, eIF4F.

The TrkB internal ribosome entry site (IRES) is an RNA element which is present in the 5' UTR sequence of the mRNA. TrkB is a neurotrophin receptor which is essential for the development and maintenance of the nervous system. The internal ribosome entry site IRES element allows cap-independent translation of TrkB which may be needed for efficient translation in neuronal dendrites.

This family represents the vascular endothelial growth factor (VEGF) internal ribosome entry site (IRES) A. VEGF is an endothelial cell mitogen with many crucial functions such as embryogenic development and wound healing. The 5' UTR of VEGF mRNA contains two IRES elements which are able to promote efficient translation at the AUG start codon, this family represents IRES A.
A ribosome binding site, or ribosomal binding site (RBS), is a sequence of nucleotides upstream of the start codon of an mRNA transcript that is responsible for the recruitment of a ribosome during the initiation of protein translation. Mostly, RBS refers to bacterial sequences, although internal ribosome entry sites (IRES) have been described in mRNAs of eukaryotic cells or viruses that infect eukaryotes. Ribosome recruitment in eukaryotes is generally mediated by the 5' cap present on eukaryotic mRNAs.
Eukaryotic translation initiation factor 4 G (eIF4G) is a protein involved in eukaryotic translation initiation and is a component of the eIF4F cap-binding complex. Orthologs of eIF4G have been studied in multiple species, including humans, yeast, and wheat. However, eIF4G is exclusively found in domain Eukarya, and not in domains Bacteria or Archaea, which do not have capped mRNA. As such, eIF4G structure and function may vary between species, although the human eIF4G 1 has been the focus of extensive studies.

Red clover necrotic mosaic virus (RCNMV) contains several structural elements present within the 3' and 5' untranslated regions (UTR) of the genome that enhance translation. In eukaryotes transcription is a prerequisite for translation. During transcription the pre-mRNA transcript is processes where a 5' cap is attached onto mRNA and this 5' cap allows for ribosome assembly onto the mRNA as it acts as a binding site for the eukaryotic initiation factor eIF4F. Once eIF4F is bound to the mRNA this protein complex interacts with the poly(A) binding protein which is present within the 3' UTR and results in mRNA circularization. This multiprotein-mRNA complex then recruits the ribosome subunits and scans the mRNA until it reaches the start codon. Transcription of viral genomes differs from eukaryotes as viral genomes produce mRNA transcripts that lack a 5’ cap site. Despite lacking a cap site viral genes contain a structural element within the 5’ UTR known as an internal ribosome entry site (IRES). IRES is a structural element that recruits the 40s ribosome subunit to the mRNA within close proximity of the start codon.
Cripavirus is a genus of viruses in the order Picornavirales, in the family Dicistroviridae. Invertebrates serve as natural hosts. There are currently nine species in this genus including the type species Cricket paralysis virus. Diseases associated with this genus include: DCV: increased reproductive potential; extremely pathogenic when injected with high associated mortality; CrPV: paralysis and death. These viruses can produce proteins directly from their RNA genome upon entering a cell; and therefore, does not require an RNA polymerase packaged in with it, as this may be produced from the genome after entering the cell. The name of the cripavirus family originates from its most famous member the Cricket Paralysis Virus. Which was made famous by its rather unusual IRES : the Cripavirus IRES. The Cripavirus IRES is an RNA element that allows the virus to bind the ribosome and translate without a need for any initiation factors – as initiation is the most regulated step of translation this allows the virus to avoid many mechanisms to inhibit viral activity.













