Bacterial translation is the process by which messenger RNA is translated into proteins in bacteria.
Eukaryotic translation is the biological process by which messenger RNA is translated into proteins in eukaryotes. It consists of four phases: initiation, elongation, termination, and recapping.
The Kozak consensus sequence is a nucleic acid motif that functions as the protein translation initiation site in most eukaryotic mRNA transcripts. Regarded as the optimum sequence for initiating translation in eukaryotes, the sequence is an integral aspect of protein regulation and overall cellular health as well as having implications in human disease. It ensures that a protein is correctly translated from the genetic message, mediating ribosome assembly and translation initiation. A wrong start site can result in non-functional proteins. As it has become more studied, expansions of the nucleotide sequence, bases of importance, and notable exceptions have arisen. The sequence was named after the scientist who discovered it, Marilyn Kozak. Kozak discovered the sequence through a detailed analysis of DNA genomic sequences.
In molecular biology, initiation factors are proteins that bind to the small subunit of the ribosome during the initiation of translation, a part of protein biosynthesis.
A bacterial initiation factor (IF) is a protein that stabilizes the initiation complex for polypeptide translation.
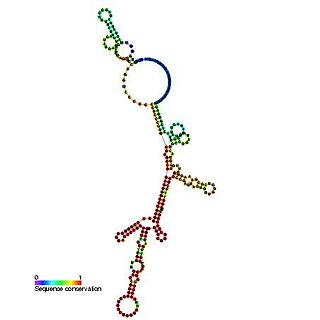
The Hepatitis C virus internal ribosome entry site, or HCV IRES, is an RNA structure within the 5'UTR of the HCV genome that mediates cap-independent translation initiation.
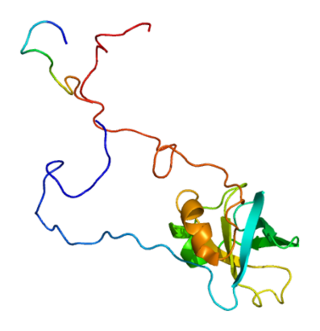
Eukaryotic translation initiation factor 1A, X-chromosomal (eIF1A) is a protein that in humans is encoded by the EIF1AX gene. This gene encodes an essential eukaryotic translation initiation factor. The protein is a component of the 43S pre-initiation complex (PIC), which mediates the recruitment of the small 40S ribosomal subunit to the 5' cap of messenger RNAs.

Eukaryotic translation initiation factor 5B is a protein that in humans is encoded by the EIF5B gene.

Eukaryotic translation initiation factor 1 (eIF1) is a protein that in humans is encoded by the EIF1 gene. It is related to yeast SUI1.
The eukaryotic small ribosomal subunit (40S) is the smaller subunit of the eukaryotic 80S ribosomes, with the other major component being the large ribosomal subunit (60S). The "40S" and "60S" names originate from the convention that ribosomal particles are denoted according to their sedimentation coefficients in Svedberg units. It is structurally and functionally related to the 30S subunit of 70S prokaryotic ribosomes. However, the 40S subunit is much larger than the prokaryotic 30S subunit and contains many additional protein segments, as well as rRNA expansion segments.
Eukaryotic translation initiation factor 4 G (eIF4G) is a protein involved in eukaryotic translation initiation and is a component of the eIF4F cap-binding complex. Orthologs of eIF4G have been studied in multiple species, including humans, yeast, and wheat. However, eIF4G is exclusively found in domain Eukarya, and not in domains Bacteria or Archaea, which do not have capped mRNA. As such, eIF4G structure and function may vary between species, although the human EIF4G1 has been the focus of extensive studies.
Eukaryotic Initiation Factor 2 (eIF2) is an eukaryotic initiation factor. It is required for most forms of eukaryotic translation initiation. eIF2 mediates the binding of tRNAiMet to the ribosome in a GTP-dependent manner. eIF2 is a heterotrimer consisting of an alpha, a beta, and a gamma subunit.
The eukaryotic initiation factor-4A (eIF4A) family consists of 3 closely related proteins EIF4A1, EIF4A2, and EIF4A3. These factors are required for the binding of mRNA to 40S ribosomal subunits. In addition these proteins are helicases that function to unwind double-stranded RNA.
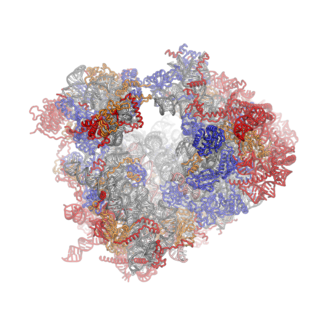
Ribosomes are a large and complex molecular machine that catalyzes the synthesis of proteins, referred to as translation. The ribosome selects aminoacylated transfer RNAs (tRNAs) based on the sequence of a protein-encoding messenger RNA (mRNA) and covalently links the amino acids into a polypeptide chain. Ribosomes from all organisms share a highly conserved catalytic center. However, the ribosomes of eukaryotes are much larger than prokaryotic ribosomes and subject to more complex regulation and biogenesis pathways. Eukaryotic ribosomes are also known as 80S ribosomes, referring to their sedimentation coefficients in Svedberg units, because they sediment faster than the prokaryotic (70S) ribosomes. Eukaryotic ribosomes have two unequal subunits, designated small subunit (40S) and large subunit (60S) according to their sedimentation coefficients. Both subunits contain dozens of ribosomal proteins arranged on a scaffold composed of ribosomal RNA (rRNA). The small subunit monitors the complementarity between tRNA anticodon and mRNA, while the large subunit catalyzes peptide bond formation.
Translational regulation refers to the control of the levels of protein synthesized from its mRNA. This regulation is vastly important to the cellular response to stressors, growth cues, and differentiation. In comparison to transcriptional regulation, it results in much more immediate cellular adjustment through direct regulation of protein concentration. The corresponding mechanisms are primarily targeted on the control of ribosome recruitment on the initiation codon, but can also involve modulation of peptide elongation, termination of protein synthesis, or ribosome biogenesis. While these general concepts are widely conserved, some of the finer details in this sort of regulation have been proven to differ between prokaryotic and eukaryotic organisms.

Eukaryotic initiation factor 3 (eIF3) is a multiprotein complex that functions during the initiation phase of eukaryotic translation. It is essential for most forms of cap-dependent and cap-independent translation initiation. In humans, eIF3 consists of 13 nonidentical subunits (eIF3a-m) with a combined molecular weight of ~800 kDa, making it the largest translation initiation factor. The eIF3 complex is broadly conserved across eukaryotes, but the conservation of individual subunits varies across organisms. For instance, while most mammalian eIF3 complexes are composed of 13 subunits, budding yeast's eIF3 has only six subunits.

Eukaryotic initiation factor 4F (eIF4F) is a heterotrimeric protein complex that binds the 5' cap of messenger RNAs (mRNAs) to promote eukaryotic translation initiation. The eIF4F complex is composed of three non-identical subunits: the DEAD-box RNA helicase eIF4A, the cap-binding protein eIF4E, and the large "scaffold" protein eIF4G. The mammalian eIF4F complex was first described in 1983, and has been a major area of study into the molecular mechanisms of cap-dependent translation initiation ever since.
The 43S preinitiation complex is a ribonucleoprotein complex that exists during an early step of eukaryotic translation initiation. The 43S PIC contains the small ribosomal subunit (40S) bound by the initiation factors eIF1, eIF1A, eIF3, and the eIF2-Met-tRNAiMet-GTP ternary complex (eIF2-TC).

DExH-box helicase 29 (DHX29) is a 155 kDa protein that in humans is encoded by the DHX29 gene.
Archaeal initiation factors are proteins that are used during the translation step of protein synthesis in archaea. The principal functions these proteins perform include ribosome RNA/mRNA recognition, delivery of the initiator Met-tRNAiMet, methionine bound tRNAi, to the 40s ribosome, and proofreading of the initiation complex.







