
Ribosomes are macromolecular machines, found within all living cells, that perform biological protein synthesis. Ribosomes link amino acids together in the order specified by the codons of messenger RNA (mRNA) molecules to form polypeptide chains. Ribosomes consist of two major components: the small and large ribosomal subunits. Each subunit consists of one or more ribosomal RNA (rRNA) molecules and many ribosomal proteins. The ribosomes and associated molecules are also known as the translational apparatus.

In molecular biology, RNA polymerase, is an enzyme that synthesizes RNA from a DNA template.
A sigma factor is a protein needed for initiation of transcription in bacteria. It is a bacterial transcription initiation factor that enables specific binding of RNA polymerase (RNAP) to gene promoters. It is homologous to archaeal transcription factor B and to eukaryotic factor TFIIB. The specific sigma factor used to initiate transcription of a given gene will vary, depending on the gene and on the environmental signals needed to initiate transcription of that gene. Selection of promoters by RNA polymerase is dependent on the sigma factor that associates with it. They are also found in plant chloroplasts as a part of the bacteria-like plastid-encoded polymerase (PEP).
Bacterial translation is the process by which messenger RNA is translated into proteins in bacteria.
Eukaryotic translation is the biological process by which transfer RNA is translated into proteins in eukaryotes. It consists of four phases: gene regulation, elongation, termination, and recycling.
Initiation factors are proteins that bind to the small subunit of the ribosome during the initiation of translation, a part of protein biosynthesis.
Eukaryotic initiation factors (eIFs) are proteins or protein complexes involved in the initiation phase of eukaryotic translation. These proteins help stabilize the formation of ribosomal preinitiation complexes around the start codon and are an important input for post-transcription gene regulation. Several initiation factors form a complex with the small 40S ribosomal subunit and Met-tRNAiMet called the 43S preinitiation complex. Additional factors of the eIF4F complex recruit the 43S PIC to the five-prime cap structure of the mRNA, from which the 43S particle scans 5'-->3' along the mRNA to reach an AUG start codon. Recognition of the start codon by the Met-tRNAiMet promotes gated phosphate and eIF1 release to form the 48S preinitiation complex, followed by large 60S ribosomal subunit recruitment to form the 80S ribosome. There exist many more eukaryotic initiation factors than prokaryotic initiation factors, reflecting the greater biological complexity of eukaryotic translation. There are at least twelve eukaryotic initiation factors, composed of many more polypeptides, and these are described below.
All bacteria require the use of three initiation factors: IF1, and IF2, for translation. Some phyla require an additional IF3.
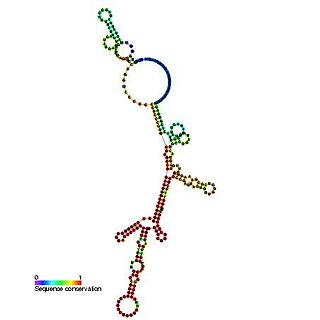
The Hepatitis C virus internal ribosome entry site, or HCV IRES, is an RNA structure within the 5'UTR of the HCV genome that mediates cap-independent translation initiation.
A ribosome binding site, or ribosomal binding site (RBS), is a sequence of nucleotides upstream of the start codon of an mRNA transcript that is responsible for the recruitment of a ribosome during the initiation of translation. Mostly, RBS refers to bacterial sequences, although internal ribosome entry sites (IRES) have been described in mRNAs of eukaryotic cells or viruses that infect eukaryotes. Ribosome recruitment in eukaryotes is generally mediated by the 5' cap present on eukaryotic mRNAs.

The prokaryotic small ribosomal subunit, or 30S subunit, is the smaller subunit of the 70S ribosome found in prokaryotes. It is a complex of the 16S ribosomal RNA (rRNA) and 19 proteins. This complex is implicated in the binding of transfer RNA to messenger RNA (mRNA). The small subunit is responsible for the binding and the reading of the mRNA during translation. The small subunit, both the rRNA and its proteins, complexes with the large 50S subunit to form the 70S prokaryotic ribosome in prokaryotic cells. This 70S ribosome is then used to translate mRNA into proteins.

EF-G is a prokaryotic elongation factor involved in protein translation. As a GTPase, EF-G catalyzes the movement (translocation) of transfer RNA (tRNA) and messenger RNA (mRNA) through the ribosome.
The eukaryotic small ribosomal subunit (40S) is the smaller subunit of the eukaryotic 80S ribosomes, with the other major component being the large ribosomal subunit (60S). The "40S" and "60S" names originate from the convention that ribosomal particles are denoted according to their sedimentation coefficients in Svedberg units. It is structurally and functionally related to the 30S subunit of 70S prokaryotic ribosomes. However, the 40S subunit is much larger than the prokaryotic 30S subunit and contains many additional protein segments, as well as rRNA expansion segments.

A protein synthesis inhibitor is a compound that stops or slows the growth or proliferation of cells by disrupting the processes that lead directly to the generation of new proteins.
Bacterial initiation factor-2 is a bacterial initiation factor.

MALSU1 is a gene on chromosome 7 in humans that encodes the protein MALSU1. This protein localizes to mitochondria and is probably involved in mitochondrial translation or the biogenesis of the large subunit of the mitochondrial ribosome.
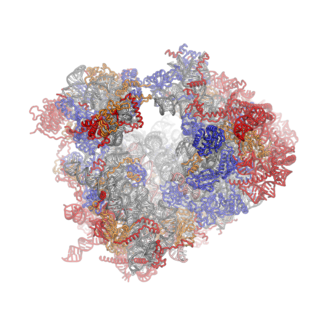
Ribosomes are a large and complex molecular machine that catalyzes the synthesis of proteins, referred to as translation. The ribosome selects aminoacylated transfer RNAs (tRNAs) based on the sequence of a protein-encoding messenger RNA (mRNA) and covalently links the amino acids into a polypeptide chain. Ribosomes from all organisms share a highly conserved catalytic center. However, the ribosomes of eukaryotes are much larger than prokaryotic ribosomes and subject to more complex regulation and biogenesis pathways. Eukaryotic ribosomes are also known as 80S ribosomes, referring to their sedimentation coefficients in Svedberg units, because they sediment faster than the prokaryotic (70S) ribosomes. Eukaryotic ribosomes have two unequal subunits, designated small subunit (40S) and large subunit (60S) according to their sedimentation coefficients. Both subunits contain dozens of ribosomal proteins arranged on a scaffold composed of ribosomal RNA (rRNA). The small subunit monitors the complementarity between tRNA anticodon and mRNA, while the large subunit catalyzes peptide bond formation.

In molecular biology, the CRM domain is an approximately 100-amino acid RNA-binding domain. The name CRM has been suggested to reflect the functions established for four characterised members of the family: Zea mays (Maize) CRS1, CAF1 and CAF2 proteins and the Escherichia coli protein YhbY. Proteins containing the CRM domain are found in eubacteria, archaea, and plants. The CRM domain is represented as a stand-alone protein in archaea and bacteria, and in single- and multi-domain proteins in plants. It has been suggested that prokaryotic CRM proteins existed as ribosome-associated proteins prior to the divergence of archaea and bacteria, and that they were co-opted in the plant lineage as RNA binding modules by incorporation into diverse protein contexts. Plant CRM domains are predicted to reside not only in the chloroplast, but also in the mitochondrion and the nucleo/cytoplasmic compartment. The diversity of the CRM domain family in plants suggests a diverse set of RNA targets.

EF-P is an essential protein that in eubacteria stimulates the formation of the first peptide bonds in protein synthesis. Studies show that EF-P prevents ribosomes from stalling during the synthesis of proteins containing consecutive prolines. EF-P binds to a site located between the binding site for the peptidyl tRNA and the exiting tRNA. It spans both ribosomal subunits with its amino-terminal domain positioned adjacent to the aminoacyl acceptor stem and its carboxyl-terminal domain positioned next to the anticodon stem-loop of the P site-bound initiator tRNA. The EF-P protein shape and size is very similar to a tRNA and interacts with the ribosome via the exit “E” site on the 30S subunit and the peptidyl-transferase center (PTC) of the 50S subunit. EF-P is a translation aspect of an unknown function, therefore It probably functions indirectly by altering the affinity of the ribosome for aminoacyl-tRNA, thus increasing their reactivity as acceptors for peptidyl transferase.
Archaeal initiation factors are proteins that are used during the translation step of protein synthesis in archaea. The principal functions these proteins perform include ribosome RNA/mRNA recognition, delivery of the initiator Met-tRNAiMet, methionine bound tRNAi, to the 40s ribosome, and proofreading of the initiation complex.











