Coxsackie B4 virus are enteroviruses that belong to the Picornaviridae family. These viruses can be found worldwide. They are positive-sense, single-stranded, non-enveloped RNA viruses with icosahedral geometry. Coxsackieviruses have two groups, A and B, each associated with different diseases. Coxsackievirus group A is known for causing hand-foot-and-mouth diseases while Group B, which contains six serotypes, can cause a varying range of symptoms like gastrointestinal distress myocarditis. Coxsackievirus B4 has a cell tropism for natural killer cells and pancreatic islet cells. Infection can lead to beta cell apoptosis which increases the risk of insulitis.

Picornaviruses are a group of related nonenveloped RNA viruses which infect vertebrates including fish, mammals, and birds. They are viruses that represent a large family of small, positive-sense, single-stranded RNA viruses with a 30 nm icosahedral capsid. The viruses in this family can cause a range of diseases including the common cold, poliomyelitis, meningitis, hepatitis, and paralysis.
The 5′ untranslated region is the region of a messenger RNA (mRNA) that is directly upstream from the initiation codon. This region is important for the regulation of translation of a transcript by differing mechanisms in viruses, prokaryotes and eukaryotes. While called untranslated, the 5′ UTR or a portion of it is sometimes translated into a protein product. This product can then regulate the translation of the main coding sequence of the mRNA. In many organisms, however, the 5′ UTR is completely untranslated, instead forming a complex secondary structure to regulate translation.
An internal ribosome entry site, abbreviated IRES, is an RNA element that allows for translation initiation in a cap-independent manner, as part of the greater process of protein synthesis. In eukaryotic translation, initiation typically occurs at the 5' end of mRNA molecules, since 5' cap recognition is required for the assembly of the initiation complex. The location for IRES elements is often in the 5'UTR, but can also occur elsewhere in mRNAs.

Aphthovirus is a viral genus of the family Picornaviridae. Aphthoviruses infect split-hooved animals, and include the causative agent of foot-and-mouth disease, Foot-and-mouth disease virus (FMDV). There are seven FMDV serotypes: A, O, C, SAT 1, SAT 2, SAT 3 and Asia 1, and four non-FMDV serotypes belonging to three additional species Bovine rhinitis A virus (BRAV), Bovine rhinitis B virus (BRBV) and Equine rhinitis A virus (ERAV).

This family represents the internal ribosome entry site (IRES) of the Picornaviruses. IRES elements allow cap and end-independent translation of mRNA in the host cell. The IRES achieves this by mediating the internal initiation of translation by recruiting a ribosomal 43S pre-initiation complex directly to the initiation codon and eliminates the requirement for the eukaryotic initiation factor eIF4F.

The bag-1 internal ribosome entry site (IRES) is a cis-acting element located in the 5 ' untranslated region of the BAG-1 protein mRNA. Its effects apoptosis through IRES mediated translation of the BAG-1 protein.

The Cripavirus internal ribosome entry site is an RNA element required for the production of capsid proteins through ribosome recruitment to an intergenic region IRES.
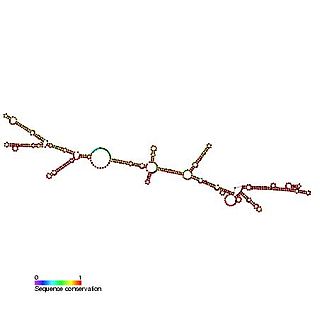
The c-sis internal ribosome entry site (IRES) is a RNA element found in the 5' UTR of the PDGF beta chain gene. The internal ribosome entry site contains three modules that can individually mediate internal ribosome entry. However, the full length sequence is required for maximal IRES activity. It is thought that the three IRES elements are somehow responsive to cellular changes and act to regulate the level of translation.
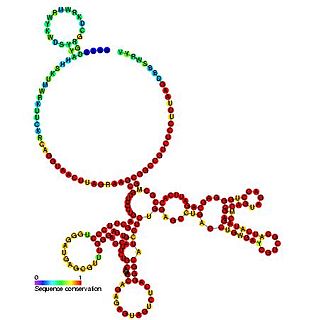
The Epstein–Barr virus nuclear-antigen internal ribosome entry site is an internal ribosome entry site (IRES) that is found in an exon in the 5' untranslated region of the Epstein–Barr virus nuclear antigen 1 (EBNA1) gene. The EBNA IRES allows EBNA1 translation to occur under situations where initiation from the 5' cap structure and ribosome scanning is reduced. It is thought that the EBNA IRES is necessary for the regulation of latent-gene expression.
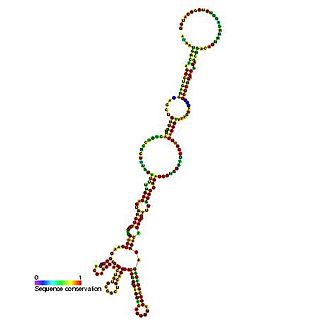
The heat shock protein 70 (Hsp70) internal ribosome entry site (IRES) is an RNA element that allows cap independent translation during conditions such as heat shock and stress. It has been shown that the 216 nucleotide long 5' UTR contains internal ribosome entry site activity.

This family represents the internal ribosome entry site (IRES) of the hepatitis A virus. HAV IRES is a 450 nucleotide long sequence located in the 735 nt long 5’ UTR of Hepatitis A viral RNA genome. IRES elements allow cap and end-independent translation of mRNA in the host cell. The IRES achieves this by mediating the internal initiation of translation by recruiting a ribosomal 40S pre-initiation complex directly to the initiation codon and eliminates the requirement for eukaryotic initiation factor, eIF4F.
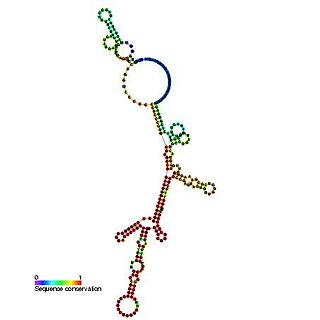
The Hepatitis C virus internal ribosome entry site, or HCV IRES, is an RNA structure within the 5'UTR of the HCV genome that mediates cap-independent translation initiation.

The Mnt internal ribosome entry site (IRES) is an RNA element. Mnt is a transcriptional repressor related to the Myc/Mad family of transcription factors. It is thought that this IRES allows efficient Mnt synthesis when cap-dependent translation initiation is reduced.

This family represents the internal ribosome entry site (IRES) of the pestiviruses. The pestivirus IRES allows cap and end-independent translation of mRNA in the host cell. The IRES achieves this by mediating the internal initiation of translation by recruiting a ribosomal 43S pre-initiation complex directly to the initiation codon and eliminates the requirement for the eukaryotic initiation factor, eIF4F. The classical swine fever virus UTR described appears to be longer at the 5' end than other pestivirus UTRs. This family represents the conserved core.
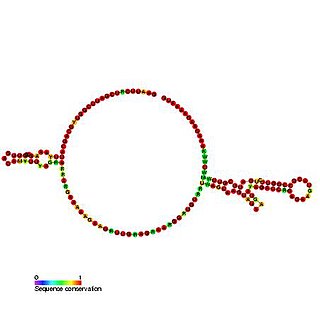
The Tobamovirus internal ribosome entry site (IRES) is an element that allows cap and end-independent translation of mRNA in the host cell. The IRES achieves this by mediating the internal initiation of translation by recruiting a ribosomal 43S pre-initiation complex directly to the initiation codon and eliminates the requirement for the eukaryotic initiation factor, eIF4F.

Senecavirus is a genus of viruses in the order Picornavirales, in the family Picornaviridae. Pig and maybe also cow serve as natural hosts. There is only one species in this genus: Senecavirus A. Senecavirus is a replication-competent oncolytic picornavirus. It has selective tropism for cancers with neuroendocrine features including small cell lung cancer (SCLC) and several pediatric solid tumors including retinoblastoma, neuroblastoma, and medulloblastoma. A Phase I clinical trial of Senecavirus in adults with neuroendocrine tumors showed that senecavirus is apparently safe to administer at doses up to 1E11 vp/kg. It has potential antineoplastic activity.

Red clover necrotic mosaic virus (RCNMV) contains several structural elements present within the 3′ and 5′ untranslated regions (UTR) of the genome that enhance translation. In eukaryotes transcription is a prerequisite for translation. During transcription the pre-mRNA transcript is processes where a 5′ cap is attached onto mRNA and this 5′ cap allows for ribosome assembly onto the mRNA as it acts as a binding site for the eukaryotic initiation factor eIF4F. Once eIF4F is bound to the mRNA this protein complex interacts with the poly(A) binding protein which is present within the 3′ UTR and results in mRNA circularization. This multiprotein-mRNA complex then recruits the ribosome subunits and scans the mRNA until it reaches the start codon. Transcription of viral genomes differs from eukaryotes as viral genomes produce mRNA transcripts that lack a 5’ cap site. Despite lacking a cap site viral genes contain a structural element within the 5’ UTR known as an internal ribosome entry site (IRES). IRES is a structural element that recruits the 40s ribosome subunit to the mRNA within close proximity of the start codon.

Picornain 3C is a protease found in picornaviruses, which cleaves peptide bonds of non-terminal sequences. Picornain 3C’s endopeptidase activity is primarily responsible for the catalytic process of selectively cleaving Gln-Gly bonds in the polyprotein of poliovirus and with substitution of Glu for Gln, and Ser or Thr for Gly in other picornaviruses. Picornain 3C are cysteine proteases related by amino acid sequence to trypsin-like serine proteases. Picornain 3C is encoded by enteroviruses, rhinoviruses, aphtoviruses and cardioviruses. These genera of picoviruses cause a wide range of infections in humans and mammals.
In molecular biology, the Nrf2 internal ribosome entry site (IRES) is an RNA element present in the 5′ UTR of the mRNA encoding the transcription factor Nrf2. It contains a stem-loop structure upstream of a ribosome binding site. This stem loop inhibits ribosome binding and translation of Nrf2 under normal conditions, but allows translation under oxidative stress conditions.
















