
In genetics, a promoter is a sequence of DNA to which proteins bind that initiate transcription of a single RNA from the DNA downstream of it. This RNA may encode a protein, or can have a function in and of itself, such as tRNA, mRNA, or rRNA. Promoters are located near the transcription start sites of genes, upstream on the DNA . Promoters can be about 100–1000 base pairs long, the sequence of which is highly dependent on the gene and product of transcription, type or class of RNA polymerase recruited to the site and species of organism.

Salmonella is a genus of rod-shaped (bacillus) Gram-negative bacteria of the family Enterobacteriaceae. The two species of Salmonella are Salmonella enterica and Salmonella bongori. S. enterica is the type species and is further divided into six subspecies that include over 2,600 serotypes. Salmonella was named after Daniel Elmer Salmon (1850–1914), an American veterinary surgeon.
In biology, quorum sensing or quorum signalling (QS) is the ability to detect and respond to cell population density by gene regulation. As one example, QS enables bacteria to restrict the expression of specific genes to the high cell densities at which the resulting phenotypes will be most beneficial. Many species of bacteria use quorum sensing to coordinate gene expression according to the density of their local population. In a similar fashion, some social insects use quorum sensing to determine where to nest. Quorum sensing may also be useful for cancer cell communications.

Salmonella enterica is a rod-headed, flagellate, facultative anaerobic, Gram-negative bacterium and a species of the genus Salmonella. A number of its serovars are serious human pathogens.

Aspergillus fumigatus is a species of fungus in the genus Aspergillus, and is one of the most common Aspergillus species to cause disease in individuals with an immunodeficiency.

Shigella flexneri is a species of Gram-negative bacteria in the genus Shigella that can cause diarrhea in humans. Several different serogroups of Shigella are described; S. flexneri belongs to group B. S. flexneri infections can usually be treated with antibiotics, although some strains have become resistant. Less severe cases are not usually treated because they become more resistant in the future. Shigella are closely related to Escherichia coli, but can be differentiated from E.coli based on pathogenicity, physiology and serology.

Yersinia pseudotuberculosis is a Gram-negative bacterium that causes Far East scarlet-like fever in humans, who occasionally get infected zoonotically, most often through the food-borne route. Animals are also infected by Y. pseudotuberculosis. The bacterium is urease positive.
Virulence factors are cellular structures, molecules and regulatory systems that enable microbial pathogens to achieve the following:
The gene rpoS encodes the sigma factor sigma-38, a 37.8 kD protein in Escherichia coli. Sigma factors are proteins that regulate transcription in bacteria. Sigma factors can be activated in response to different environmental conditions. rpoS is transcribed in late exponential phase, and RpoS is the primary regulator of stationary phase genes. RpoS is a central regulator of the general stress response and operates in both a retroactive and a proactive manner: it not only allows the cell to survive environmental challenges, but it also prepares the cell for subsequent stresses (cross-protection). The transcriptional regulator CsgD is central to biofilm formation, controlling the expression of the curli structural and export proteins, and the diguanylate cyclase, adrA, which indirectly activates cellulose production. The rpoS gene most likely originated in the gammaproteobacteria.

The MicA RNA is a small non-coding RNA that was discovered in E. coli during a large scale screen. Expression of SraD is highly abundant in stationary phase, but low levels could be detected in exponentially growing cells as well.

Invasion gene associated RNA is a small non-coding RNA involved in regulating one of the major outer cell membrane porin proteins in Salmonella species.

An Hfq binding sRNA is an sRNA that binds the bacterial RNA binding protein called Hfq. A number of bacterial small RNAs which have been shown to bind to Hfq have been characterised . Many of these RNAs share a similar structure composed of three stem-loops. Several studies have expanded this list, and experimentally validated a total of 64 Hfq binding sRNA in Salmonella Typhimurium. A transcriptome wide study on Hfq binding sites in Salmonella mapped 126 Hfq binding sites within sRNAs. Genomic SELEX has been used to show that Hfq binding RNAs are enriched in the sequence motif 5′-AAYAAYAA-3′. Genome-wide study identified 40 candidate Hfq-dependent sRNAs in plant pathogen Erwinia amylovora. 12 of them were confirmed by Northern blot.

Plant disease resistance protects plants from pathogens in two ways: by pre-formed structures and chemicals, and by infection-induced responses of the immune system. Relative to a susceptible plant, disease resistance is the reduction of pathogen growth on or in the plant, while the term disease tolerance describes plants that exhibit little disease damage despite substantial pathogen levels. Disease outcome is determined by the three-way interaction of the pathogen, the plant and the environmental conditions.

FourU thermometers are a class of non-coding RNA thermometers found in Salmonella. They are named 'FourU' due to the four highly conserved uridine nucleotides found directly opposite the Shine-Dalgarno sequence on hairpin II (pictured). RNA thermometers such as FourU control regulation of temperature via heat shock proteins in many prokaryotes. FourU thermometers are relatively small RNA molecules, only 57 nucleotides in length, and have a simple two-hairpin structure.
Bacterial small RNAs (sRNA) are small RNAs produced by bacteria; they are 50- to 500-nucleotide non-coding RNA molecules, highly structured and containing several stem-loops. Numerous sRNAs have been identified using both computational analysis and laboratory-based techniques such as Northern blotting, microarrays and RNA-Seq in a number of bacterial species including Escherichia coli, the model pathogen Salmonella, the nitrogen-fixing alphaproteobacterium Sinorhizobium meliloti, marine cyanobacteria, Francisella tularensis, Streptococcus pyogenes, the pathogen Staphylococcus aureus, and the plant pathogen Xanthomonas oryzae pathovar oryzae. Bacterial sRNAs affect how genes are expressed within bacterial cells via interaction with mRNA or protein, and thus can affect a variety of bacterial functions like metabolism, virulence, environmental stress response, and structure.

An RNA thermometer is a temperature-sensitive non-coding RNA molecule which regulates gene expression. RNA thermometers often regulate genes required during either a heat shock or cold shock response, but have been implicated in other regulatory roles such as in pathogenicity and starvation.
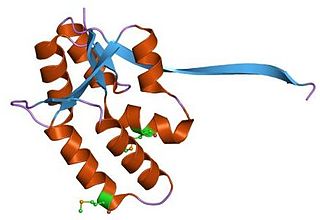
VapBC is the largest family of type II toxin-antitoxin system genetic loci in prokaryotes. VapBC operons consist of two genes: VapC encodes a toxic PilT N-terminus (PIN) domain, and VapB encodes a matching antitoxin. The toxins in this family are thought to perform RNA cleavage, which is inhibited by the co-expression of the antitoxin, in a manner analogous to a poison and antidote.
In molecular biology, the FasX small RNA (fibronectin/fibrinogen-binding/haemolytic-activity/streptokinase-regulator-X) is a non-coding small RNA (sRNA) produced by all group A Streptococcus. FasX has also been found in species of group D and group G Streptococcus. FasX regulates expression of secreted virulence factor streptokinase (SKA), encoded by the ska gene. FasX base pairs to the 5' end of the ska mRNA, increasing the stability of the mRNA, resulting in elevated levels of streptokinase expression. FasX negatively regulates the expression of pili and fibronectin-binding proteins on the bacterial cell surface. It binds to the 5' untranslated region of genes in the FCT-region in a serotype-specific manner, reducing the stability of and inhibiting translation of the pilus biosynthesis operon mRNA by occluding the ribosome-binding site through a simple Watson-Crick base-pairing mechanism.
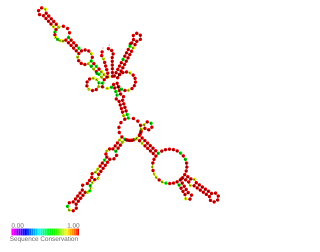
The 3′ UTR of mRNA hilD, a master regulator of Salmonella pathogenicity island 1 (SPI-1), is a prokaryotic example of functional 3'UTR. The 3'UTR is a target for hilD mRNA degradation by the degradosome and it may play a role in hilD and SPI-1 expression by serving as a target for the Hfq RNA chaperone. Under non-invasive conditions it is necessary to keep low levels of SPI-1 expression. It plays a role in S. Typhimurium virulence as a regulatory motif.

The IsrM RNA is a small non-coding RNA discovered in Salmonella pathogenicity island, which is not found in E.coli. It is important for invasion of epithelial cells, intracellular replication inside macrophages, virulence and colonisation in mice. It targets the SopA and HilE mRNAs, virulence factors essential for bacterial invasion. It is a first pathogenicity island-encoded sRNA shown to be directly involved in Salmonella pathogenesis.














