The bit is the most basic unit of information in computing and digital communications. The name is a portmanteau of binary digit. The bit represents a logical state with one of two possible values. These values are most commonly represented as either "1" or "0", but other representations such as true/false, yes/no, on/off, or +/− are also widely used.

Bioinformatics is an interdisciplinary field of science that develops methods and software tools for understanding biological data, especially when the data sets are large and complex. Bioinformatics uses biology, chemistry, physics, computer science, computer programming, information engineering, mathematics and statistics to analyze and interpret biological data. The subsequent process of analyzing and interpreting data is referred to as computational biology.

The cell is the basic structural and functional unit of all forms of life. Every cell consists of cytoplasm enclosed within a membrane, and contains many macromolecules such as proteins, DNA and RNA, as well as many small molecules of nutrients and metabolites. The term comes from the Latin word cellula meaning 'small room'.
Computing is any goal-oriented activity requiring, benefiting from, or creating computing machinery. It includes the study and experimentation of algorithmic processes, and development of both hardware and software. Computing has scientific, engineering, mathematical, technological and social aspects. Major computing disciplines include computer engineering, computer science, cybersecurity, data science, information systems, information technology, digital art and software engineering.
In communications and information processing, code is a system of rules to convert information—such as a letter, word, sound, image, or gesture—into another form, sometimes shortened or secret, for communication through a communication channel or storage in a storage medium. An early example is an invention of language, which enabled a person, through speech, to communicate what they thought, saw, heard, or felt to others. But speech limits the range of communication to the distance a voice can carry and limits the audience to those present when the speech is uttered. The invention of writing, which converted spoken language into visual symbols, extended the range of communication across space and time.

Computer data storage is a technology consisting of computer components and recording media that are used to retain digital data. It is a core function and fundamental component of computers.
Data storage is the recording (storing) of information (data) in a storage medium. Handwriting, phonographic recording, magnetic tape, and optical discs are all examples of storage media. Biological molecules such as RNA and DNA are considered by some as data storage. Recording may be accomplished with virtually any form of energy. Electronic data storage requires electrical power to store and retrieve data.
A stored-program computer is a computer that stores program instructions in electronically or optically accessible memory. This contrasts with systems that stored the program instructions with plugboards or similar mechanisms.

Computational biology refers to the use of data analysis, mathematical modeling and computational simulations to understand biological systems and relationships. An intersection of computer science, biology, and big data, the field also has foundations in applied mathematics, chemistry, and genetics. It differs from biological computing, a subfield of computer engineering which uses bioengineering to build computers.

Sharing is the joint use of a resource or space. It is also the process of dividing and distributing. In its narrow sense, it refers to joint or alternating use of inherently finite goods, such as a common pasture or a shared residence. Still more loosely, "sharing" can actually mean giving something as an outright gift: for example, to "share" one's food really means to give some of it as a gift. Sharing is a basic component of human interaction, and is responsible for strengthening social ties and ensuring a person’s well-being.
The floating-gate MOSFET (FGMOS), also known as a floating-gate MOS transistor or floating-gate transistor, is a type of metal–oxide–semiconductor field-effect transistor (MOSFET) where the gate is electrically isolated, creating a floating node in direct current, and a number of secondary gates or inputs are deposited above the floating gate (FG) and are electrically isolated from it. These inputs are only capacitively connected to the FG. Since the FG is surrounded by highly resistive material, the charge contained in it remains unchanged for long periods of time, nowadays typically longer than 10 years. Usually Fowler-Nordheim tunneling and hot-carrier injection mechanisms are used to modify the amount of charge stored in the FG.
Data format management (DFM) is the application of a systematic approach to the selection and use of the data formats used to encode information for storage on a computer.
A file format is a standard way that information is encoded for storage in a computer file. It specifies how bits are used to encode information in a digital storage medium. File formats may be either proprietary or free.

Plant genetics is the study of genes, genetic variation, and heredity specifically in plants. It is generally considered a field of biology and botany, but intersects frequently with many other life sciences and is strongly linked with the study of information systems. Plant genetics is similar in many ways to animal genetics but differs in a few key areas.
Natural computing, also called natural computation, is a terminology introduced to encompass three classes of methods: 1) those that take inspiration from nature for the development of novel problem-solving techniques; 2) those that are based on the use of computers to synthesize natural phenomena; and 3) those that employ natural materials to compute. The main fields of research that compose these three branches are artificial neural networks, evolutionary algorithms, swarm intelligence, artificial immune systems, fractal geometry, artificial life, DNA computing, and quantum computing, among others.

Artificial life is a field of study wherein researchers examine systems related to natural life, its processes, and its evolution, through the use of simulations with computer models, robotics, and biochemistry. The discipline was named by Christopher Langton, an American theoretical biologist, in 1986. In 1987 Langton organized the first conference on the field, in Los Alamos, New Mexico. There are three main kinds of alife, named for their approaches: soft, from software; hard, from hardware; and wet, from biochemistry. Artificial life researchers study traditional biology by trying to recreate aspects of biological phenomena.
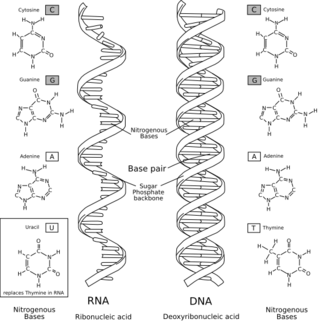
DNA digital data storage is the process of encoding and decoding binary data to and from synthesized strands of DNA.
An inforg is an informationally embodied organism, entity made up of information, that exists in the infosphere. These informationally embodied organisms are also called natural agents.
Nicholas Goldman is a group leader and senior scientist at the European Bioinformatics Institute (EBI), located on the Wellcome Genome Campus in Hinxton, Cambridgeshire, England. He began working at the EBI in 2002, and became a senior scientist there in 2009. His group's research focuses on evolutionary genetics and genomics. He and his EBI colleague Ewan Birney, along with other researchers, developed a tool for DNA digital data storage, on which they successfully encoded all the sonnets of William Shakespeare, Martin Luther King Jr.'s 1963 "I Have a Dream" speech, a PDF of the 1953 paper "Molecular Structure of Nucleic Acids: A Structure for Deoxyribose Nucleic Acid", and a photo of their own institute. They described their results in a 2013 paper in Nature.








