
Sindbis virus (SINV) is a member of the Togaviridae family, in the alphavirus subfamily. The virus was first isolated in 1952 in Cairo, Egypt. The virus is transmitted by mosquitoes SINV causes sindbis fever in humans and the symptoms include arthralgia, rash and malaise. Sindbis fever is most common in South and East Africa, Egypt, Israel, Philippines and parts of Australia. Sindbis virus is an "arbovirus" (arthropod-borne) and is maintained in nature by transmission between vertebrate (bird) hosts and invertebrate (mosquito) vectors. Humans are infected with Sindbis virus when bitten by an infected mosquito. SINV has been linked to Pogosta disease in Finland, Ockelbo disease in Sweden and Karelian fever in Russia.
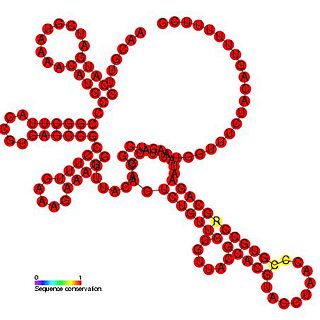
The Bamboo mosaic potexvirus (BaMV) cis-regulatory element represents a cloverleaf-like cis-regulatory element found in the 3' UTR of the bamboo mosaic virus. This family is thought to play an important role in the initiation of minus-strand RNA synthesis and may also be involved in the regulation of viral replication.
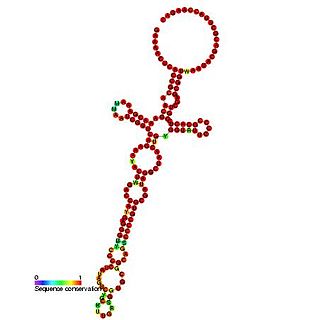
The bamboo mosaic virus satellite RNA cis-regulatory element is an RNA element found in the 5' UTR of the genome of the bamboo mosaic virus. This element is thought to be essential for efficient RNA replication.

The Citrus tristeza virus replication signal is a regulatory element involved in a viral replication signal which is highly conserved in citrus tristeza viruses. Replication signals are required for viral replication and are usually found near the 5' and 3' termini of protein coding genes. This element is predicted to form ten stem loop structures some of which are essential for functions that provide for efficient viral replication.
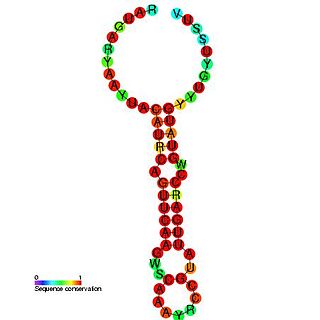
Enterovirus cis-acting replication element is a small RNA hairpin in the coding region of protein 2C as the site in PV1(M) RNA that is used as the primary template for the in vitro uridylylation. The first step in the replication of the plus-stranded poliovirus RNA is the synthesis of a complementary minus strand. This process is initiated by the covalent attachment of uridine monophosphate (UMP) to the terminal protein VPg, yielding VPgpU and VPgpUpU.

The Hepatitis C virus (HCV) cis-acting replication element (CRE) is an RNA element which is found in the coding region of the RNA-dependent RNA polymerase NS5B. Mutations in this family have been found to cause a blockage in RNA replication and it is thought that both the primary sequence and the structure of this element are crucial for HCV RNA replication.
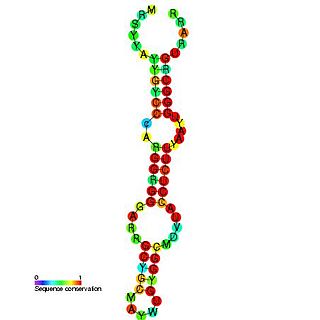
Hepatitis C virus stem-loop VII is a regulatory element found in the coding region of the RNA-dependent RNA polymerase gene, NS5B. Similarly to stem-loop IV, the stem-loop structure is important for colony formation, though its exact function and mechanism are unknown.
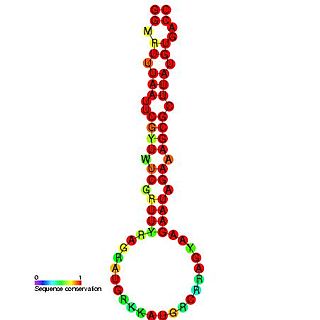
This family represents a rotavirus cis-acting replication element (CRE) found at the 3'-end of rotavirus mRNAs. The family is thought to promote the synthesis of minus strand RNA to form viral dsRNA.
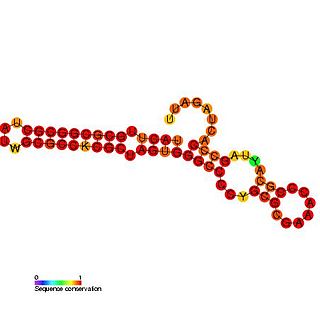
The Rubella virus 3' cis-acting element RNA family represents a cis-acting element found at the 3' UTR in the rubella virus. This family contains three conserved step loop structures. Calreticulin (CAL), which is known to bind calcium in most eukaryotic cells, is able to specifically bind to the first stem loop of this RNA. CAL binding is thought to be related to viral pathogenesis and in particular arthritis which occurs frequently in rubella infections in adults and is independent of viral viability. All stem loop structures are thought to be important for efficient viral replication and deletion of stem loop three is known to be lethal.

Human rhinovirus internal cis-acting regulatory element (CRE) is a CRE from the human rhinoviruses. The CRE is located within the genome segment encoding the capsid proteins so is found in a protein coding region. The element is essential for efficient viral replication and it has been suggested that the CRE is required for initiation of minus-strand RNA synthesis.
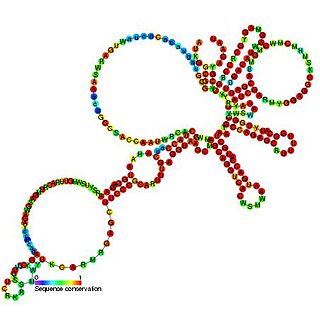
The Togavirus 5' plus strand cis-regulatory element is an RNA element which is thought to be essential for both plus and minus strand RNA synthesis.
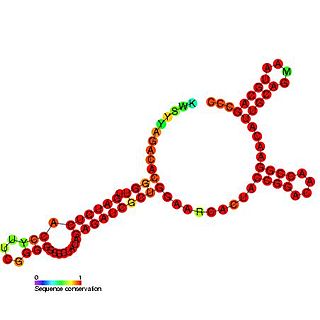
Tombusvirus 3' UTR is an important cis-regulatory region of the Tombus virus genome.
Tombusvirus 5' UTR is an important cis-regulatory region of the Tombus virus genome.
Tombus virus defective interfering (DI) RNA region 3 is an important cis-regulatory region identified in the 3' UTR of Tombusvirus defective interfering particles (DI).

The Upstream pseudoknot (UPSK) domain is an RNA element found in the turnip yellow mosaic virus, beet virus Q, barley stripe mosaic virus and tobacco mosaic virus, which is thought to be needed for efficient transcription. Disruption of the pseudoknot structure gives rise to a 50% drop in transcription efficiency. This element acts in conjunction with the Tymovirus/Pomovirus tRNA-like 3' UTR element to enhance translation.
Cis-acting replication elements bring together the 5' and 3' ends during replication of positive-sense single-stranded RNA viruses and double-stranded RNA viruses.
In molecular biology, the Norovirus cis-acting replication element (CRE) is an RNA element which is found in the coding region of the RNA-dependent RNA polymerase in Norovirus. It occurs near to the 5' end of the RNA dependant RNA polymerase gene, this is the same location that the Hepatitis A virus cis-acting replication element is found in.


















