
Alternative splicing, or alternative RNA splicing, or differential splicing, is an alternative splicing process during gene expression that allows a single gene to code for multiple proteins. In this process, particular exons of a gene may be included within or excluded from the final, processed messenger RNA (mRNA) produced from that gene. This means the exons are joined in different combinations, leading to different (alternative) mRNA strands. Consequently, the proteins translated from alternatively spliced mRNAs will contain differences in their amino acid sequence and, often, in their biological functions. Notably, alternative splicing allows the human genome to direct the synthesis of many more proteins than would be expected from its 20,000 protein-coding genes.

Poliovirus, the causative agent of polio, is a serotype of the species Enterovirus C, in the family of Picornaviridae.

Picornaviruses are a group of related nonenveloped RNA viruses which infect vertebrates including fish, mammals, and birds. They are viruses that represent a large family of small, positive-sense, single-stranded RNA viruses with a 30 nm icosahedral capsid. The viruses in this family can cause a range of diseases including the common cold, poliomyelitis, meningitis, hepatitis, and paralysis.
Polyadenylation is the addition of a poly(A) tail to an RNA transcript, typically a messenger RNA (mRNA). The poly(A) tail consists of multiple adenosine monophosphates; in other words, it is a stretch of RNA that has only adenine bases. In eukaryotes, polyadenylation is part of the process that produces mature mRNA for translation. In many bacteria, the poly(A) tail promotes degradation of the mRNA. It, therefore, forms part of the larger process of gene expression.

Antisense RNA (asRNA), also referred to as antisense transcript, natural antisense transcript (NAT) or antisense oligonucleotide, is a single stranded RNA that is complementary to a protein coding messenger RNA (mRNA) with which it hybridizes, and thereby blocks its translation into protein. asRNAs have been found in both prokaryotes and eukaryotes, and can be classified into short and long non-coding RNAs (ncRNAs). The primary function of asRNA is regulating gene expression. asRNAs may also be produced synthetically and have found wide spread use as research tools for gene knockdown. They may also have therapeutic applications.
VPg is a protein that is covalently attached to the 5′ end of positive strand viral RNA and acts as a primer during RNA synthesis in a variety of virus families including Picornaviridae, Potyviridae and Caliciviridae. There are some studies showing that a possible VPg protein is also present in astroviridae, however, experimental evidence for this has not yet been provided and requires further study. The primer activity of VPg occurs when the protein becomes uridylated, providing a free hydroxyl that can be extended by the virally encoded RNA-dependent RNA polymerase. For some virus families, VPg also has a role in translation initiation by acting like a 5' mRNA cap.

A long terminal repeat (LTR) is a pair of identical sequences of DNA, several hundred base pairs long, which occur in eukaryotic genomes on either end of a series of genes or pseudogenes that form a retrotransposon or an endogenous retrovirus or a retroviral provirus. All retroviral genomes are flanked by LTRs, while there are some retrotransposons without LTRs. Typically, an element flanked by a pair of LTRs will encode a reverse transcriptase and an integrase, allowing the element to be copied and inserted at a different location of the genome. Copies of such an LTR-flanked element can often be found hundreds or thousands of times in a genome. LTR retrotransposons comprise about 8% of the human genome.
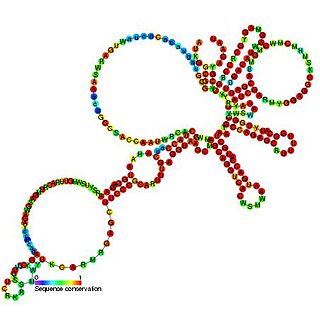
The bamboo mosaic virus satellite RNA cis-regulatory element is an RNA element found in the 5' UTR of the genome of the bamboo mosaic virus. This element is thought to be essential for efficient RNA replication.
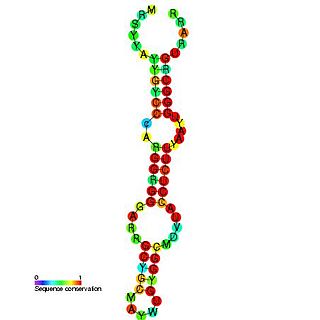
Hepatitis C virus stem-loop VII is a regulatory element found in the coding region of the RNA-dependent RNA polymerase gene, NS5B. Similarly to stem-loop IV, the stem-loop structure is important for colony formation, though its exact function and mechanism are unknown.

The hepatitis delta virus (HDV) ribozyme is a non-coding RNA found in the hepatitis delta virus that is necessary for viral replication and is the only known human virus that utilizes ribozyme activity to infect its host. The ribozyme acts to process the RNA transcripts to unit lengths in a self-cleavage reaction during replication of the hepatitis delta virus, which is thought to propagate by a double rolling circle mechanism. The ribozyme is active in vivo in the absence of any protein factors and was the fastest known naturally occurring self-cleaving RNA at the time of its discovery.
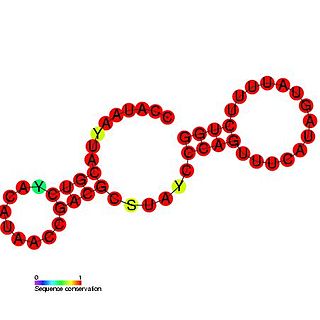
The Potato virus X cis-acting regulatory element is a cis-acting regulatory element found in the 3' UTR of the Potato virus X genome. This element has been found to be required for minus strand RNA accumulation and is essential for efficient viral replication.

The retroviral psi packaging element, also known as the Ψ RNA packaging signal, is a cis-acting RNA element identified in the genomes of the retroviruses Human immunodeficiency virus (HIV) and Simian immunodeficiency virus (SIV). It is involved in regulating the essential process of packaging the retroviral RNA genome into the viral capsid during replication. The final virion contains a dimer of two identical unspliced copies of the viral genome.
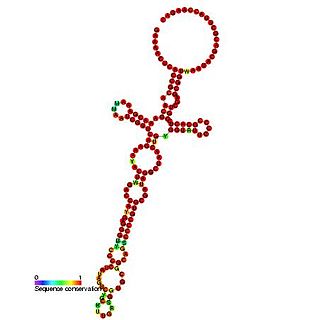
The Togavirus 5′ plus strand cis-regulatory element is an RNA element which is thought to be essential for both plus and minus strand RNA synthesis.
Trans-regulatory elements (TRE) are DNA sequences encoding upstream regulators, which may modify or regulate the expression of distant genes. Trans-acting factors interact with cis-regulatory elements to regulate gene expression. TRE mediates expression profiles of a large number of genes via trans-acting factors. While TRE mutations affect gene expression, it is also one of the main driving factors for evolutionary divergence in gene expression.
Rev is a transactivating protein that is essential to the regulation of HIV-1 protein expression. A nuclear localization signal is encoded in the rev gene, which allows the Rev protein to be localized to the nucleus, where it is involved in the export of unspliced and incompletely spliced mRNAs. In the absence of Rev, mRNAs of the HIV-1 late (structural) genes are retained in the nucleus, preventing their translation.
In molecular biology, the Hepatitis A virus cis-acting replication element (CRE) is an RNA element which is found in the coding region of the RNA-dependent RNA polymerase in Hepatitis A virus (HAV). It is larger than the CREs found in related Picornavirus species, but is thought to be functionally similar. It is thought to be involved in uridylylation of VPg.
In molecular biology, the Avian encephalitis virus cis-acting replication element (CRE) is an s an RNA element which is found in the coding region of the RNA-dependent RNA polymerase in Avian encephalitis virus (AEV). It is structurally similar to the Hepatitis A virus cis-acting replication element.
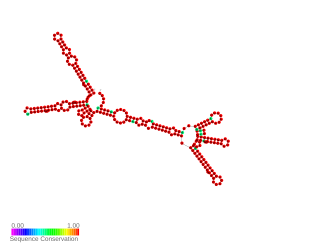
Hepatitis B virus PRE 1151–1410 is a part of 500 base pair long HBV PRE, that has been proposed to be the hepatitis B virus (HBV) RNA export element. However, the function is controversial and new regulatory elements have been predicted within PRE. PRE 1151–1410 enhances nuclear export of intronless transcripts and represses the splicing mechanism to a comparable degree to that of the full-length PRE. Hence it was proposed to be the core HBV PRE element. PRE1151–1410 contains 3 known regulatory elements: PRE SL-alpha, human La protein binding site, SRE-1.
Flavivirus 3' UTR are untranslated regions in the genome of viruses in the genus Flavivirus.












