Related Research Articles
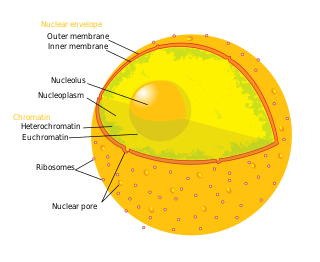
The nucleolus is the largest structure in the nucleus of eukaryotic cells. It is best known as the site of ribosome biogenesis. Nucleoli also participate in the formation of signal recognition particles and play a role in the cell's response to stress. Nucleoli are made of proteins, DNA and RNA and form around specific chromosomal regions called nucleolar organizing regions. Malfunction of nucleoli can be the cause of several human conditions called "nucleolopathies" and the nucleolus is being investigated as a target for cancer chemotherapy.
Small nuclear RNA (snRNA) is a class of small RNA molecules that are found within the splicing speckles and Cajal bodies of the cell nucleus in eukaryotic cells. The length of an average snRNA is approximately 150 nucleotides. They are transcribed by either RNA polymerase II or RNA polymerase III. Their primary function is in the processing of pre-messenger RNA (hnRNA) in the nucleus. They have also been shown to aid in the regulation of transcription factors or RNA polymerase II, and maintaining the telomeres.
In molecular biology, Small nucleolar RNAs (snoRNAs) are a class of small RNA molecules that primarily guide chemical modifications of other RNAs, mainly ribosomal RNAs, transfer RNAs and small nuclear RNAs. There are two main classes of snoRNA, the C/D box snoRNAs, which are associated with methylation, and the H/ACA box snoRNAs, which are associated with pseudouridylation. SnoRNAs are commonly referred to as guide RNAs but should not be confused with the guide RNAs that direct RNA editing in trypanosomes.
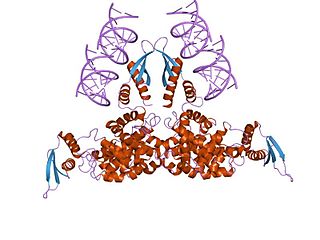
Ribonuclease III (BRENDA 3.1.26.3) is a type of ribonuclease that recognizes dsRNA and cleaves it at specific targeted locations to transform them into mature RNAs. These enzymes are a group of endoribonucleases that are characterized by their ribonuclease domain, which is labelled the RNase III domain. They are ubiquitous compounds in the cell and play a major role in pathways such as RNA precursor synthesis, RNA Silencing, and the pnp autoregulatory mechanism.

Ribosome biogenesis is the process of making ribosomes. In prokaryotes, this process takes place in the cytoplasm with the transcription of many ribosome gene operons. In eukaryotes, it takes place both in the cytoplasm and in the nucleolus. It involves the coordinated function of over 200 proteins in the synthesis and processing of the three prokaryotic or four eukaryotic rRNAs, as well as assembly of those rRNAs with the ribosomal proteins. Most of the ribosomal proteins fall into various energy-consuming enzyme families including ATP-dependent RNA helicases, AAA-ATPases, GTPases, and kinases. About 60% of a cell's energy is spent on ribosome production and maintenance.

Shq1p is a protein involved in the rRNA processing pathway. It was discovered by Pok Yang in the Chanfreau laboratory at UCLA. Depletion of Shq1p has led to decreased level of various H/ACA box snoRNAs and certain pre-rRNA intermediates.

In molecular biology, U3 snoRNA is a non-coding RNA found predominantly in the nucleolus. U3 has C/D box motifs that technically make it a member of the box C/D class of snoRNAs; however, unlike other C/D box snoRNAs, it has not been shown to direct 2'-O-methylation of other RNAs. Rather, U3 is thought to guide site-specific cleavage of ribosomal RNA (rRNA) during pre-rRNA processing.

In molecular biology, U14 small nucleolar RNA is a non-coding RNA required for early cleavages of eukaryotic precursor rRNAs. In yeasts, this molecule possess a stem-loop region which is essential for function. A similar structure, but with a different consensus sequence, is found in plants, but is absent in vertebrates. In human there are two closely related copies called SNORD14A and SNORD14B that are expressed from the intron of their host gene ribosomal protein Rps13.

In molecular biology, Small nucleolar RNA SNORA74 (U19) belongs to the H/ACA class of snoRNAs. snoRNAs bind a number of proteins to form snoRNP complexes. This class is thought to guide the sites of modification of uridines to pseudouridines by forming direct base pairing interactions with substrate RNAs. Targets may include ribosomal and spliceosomal RNAs but the exact functions of many snoRNAs, including U19, are not confirmed. Co-precipitation of U19 snoRNA with RNase MRP RNA suggests that U19 may be involved in pre-rRNA processing.
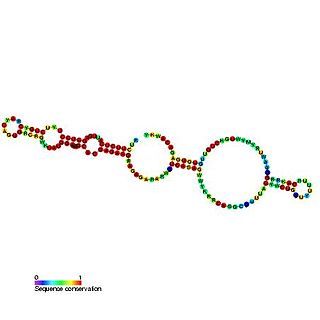
In molecular biology, U23 belongs to the H/ACA class of snoRNAs. snoRNAs bind a number of proteins to form snoRNP complexes. This class are thought to guide the sites of modification of uridines to pseudouridines by forming direct base pairing interactions with substrate RNAs. Targets include ribosomal and spliceosomal RNAs as well as the Trypanosoma spliced leader RNA as possibly other, still unknown cellular RNAs. U23 can direct the pseudouridylation of U97 in human 18S rRNA. U23 is encoded within intron 12 of the nucleolin gene in human, mouse, rat chicken, and Xenopus laevis.

In molecular biology, SNORD15 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.

In molecular biology, snoRNA U16 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.
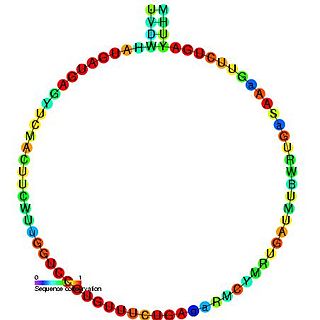
In molecular biology, SNORD18 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.

In molecular biology, Small nucleolar RNA SNORD27 is a member of the C/D class of snoRNA which contain the C (UGAUGA) and D (CUGA) box motifs. U27 is encoded within the U22 snoRNA host gene (UHG) in mammals and is thought to act as a 2'-O-ribose methylation guide for ribosomal RNA. This family also contains several related snoRNAs from yeast and plants.
In molecular biology, snoRNA U62 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.
In molecular biology, snoRNA U63 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.
In molecular biology, snoRNA U73 is a non-coding RNA (ncRNA) molecule which functions in the biogenesis (modification) of other small nuclear RNAs (snRNAs). This type of modifying RNA is located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.

In molecular biology, U8 small nucleolar RNA is the RNA component of a small RNA:protein complex which is required for biogenesis of mature large subunit ribosomal RNAs, 5.8S and 28S rRNAs.

rRNA 2'-O-methyltransferase fibrillarin is an enzyme that in humans is encoded by the FBL gene.
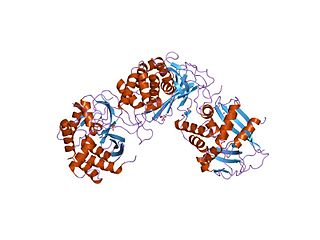
In molecular biology, Endoribonuclease XendoU refers to a protein domain. This particular entry represents endoribonucleases involved in RNA biosynthesis which has been named XendoU in Xenopus laevis. This protein domain belongs to a family of evolutionarily related proteins. XendoU is a U-specific metal dependent enzyme that produces products with a 2'-3' cyclic phosphate termini.
References
- ↑ Marmier-Gourrier N, Cle´ry A, Schlotter F, Branlant C. A second base pair interaction between U3 small nucleolar RNA and the 5’-ETS region is required for early cleavage of the yeast pre-ribosomal RNA. Nucleic Acids Research. 2011; 39.
- ↑ Gerbi SA, Borovjagin AV. Pre-Ribosomal RNA Processing in Multicellular Organisms. In: Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000-.
- ↑ Chooi WY, Leiby KR. An electron microscopic method for localization of ribosomal proteins during transcription of ribosomal DNA: A method for studying protein assembly. Proc Natl Acad Sci. 1981;78:4823–4827
- ↑ Hadjiolov AA. The Nucleolus and Ribosome Biogenesis. Vienna: Springer-Verlag KG. 1985 .
- ↑ Gerbi SA, Borovjagin AV. Pre-Ribosomal RNA Processing in Multicellular Organisms. In: Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000-.
- ↑ Borovjagin AV, Gerbi SA. U3 small nucleolar RNA is essential for cleavage at sites 1, 2, and 3 in pRNA and determines which rRNA processing pathway is taken in Xenopus oocytes. J Mol Biol. 1999;286:1347–1363
- ↑ Borovjagin AV, Gerbi SA. Xenopus U3 snoRNA docks on pre-rRNA through a novel base-pairing interaction. submitted. 2003
- ↑ Herrera A, Olson MOJ. Association of protein C23 with rapidly labeled nucleolar RNA. Biochemistry. 1986;25:6258–6264.
- ↑ Savino R, Hitti Y, Gerbi SA. Genes for Xenopus laevis U3 small nuclear RNA. Nucleic Acids Res. 1992;20:5435–5442.
- ↑ Marmier-Gourrier N, Cle´ry A, Schlotter F, Branlant C. A second base pair interaction between U3 small nucleolar RNA and the 5’-ETS region is required for early cleavage of the yeast pre-ribosomal RNA. Nucleic Acids Research. 2011; 39.
- ↑ Marmier-Gourrier N, Cle´ry A, Schlotter F, Branlant C. A second base pair interaction between U3 small nucleolar RNA and the 5’-ETS region is required for early cleavage of the yeast pre-ribosomal RNA. Nucleic Acids Research. 2011; 39.
- ↑ Gerbi SA, Borovjagin AV. Pre-Ribosomal RNA Processing in Multicellular Organisms. In: Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000-.
- ↑ Gerbi SA, Borovjagin AV. Pre-Ribosomal RNA Processing in Multicellular Organisms. In: Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000-.