Related Research Articles

Antisense RNA (asRNA), also referred to as antisense transcript, natural antisense transcript (NAT) or antisense oligonucleotide, is a single stranded RNA that is complementary to a protein coding messenger RNA (mRNA) with which it hybridizes, and thereby blocks its translation into protein. The asRNAs have been found in both prokaryotes and eukaryotes, and can be classified into short and long non-coding RNAs (ncRNAs). The primary function of asRNA is regulating gene expression. asRNAs may also be produced synthetically and have found wide spread use as research tools for gene knockdown. They may also have therapeutic applications.
In genetics, a sense strand, or coding strand, is the segment within double-stranded DNA that carries the translatable code in the 5′ to 3′ direction, and which is complementary to the antisense strand of DNA, or template strand, which does not carry the translatable code in the 5′ to 3′ direction. The sense strand is the strand of DNA that has the same sequence as the mRNA, which takes the antisense strand as its template during transcription, and eventually undergoes translation into a protein. The antisense strand is thus responsible for the RNA that is later translated to protein, while the sense strand possesses a nearly identical makeup to that of the mRNA.

N-myc proto-oncogene protein also known as N-Myc or basic helix-loop-helix protein 37 (bHLHe37), is a protein that in humans is encoded by the MYCN gene.
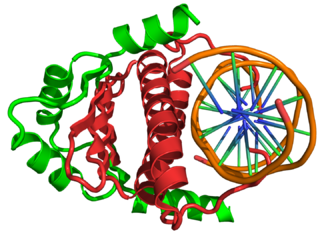
In the field of molecular biology, myocyte enhancer factor-2 (Mef2) proteins are a family of transcription factors which through control of gene expression are important regulators of cellular differentiation and consequently play a critical role in embryonic development. In adult organisms, Mef2 proteins mediate the stress response in some tissues. Mef2 proteins contain both MADS-box and Mef2 DNA-binding domains.

YY1 is a transcriptional repressor protein in humans that is encoded by the YY1 gene.

GATA3 is a transcription factor that in humans is encoded by the GATA3 gene. Studies in animal models and humans indicate that it controls the expression of a wide range of biologically and clinically important genes.

KCNQ1 overlapping transcript 1, also known as KCNQ1OT1, is a long non-coding RNA gene found in the KCNQ1 locus. This locus consists of 8–10 protein-coding genes, specifically expressed from the maternal allele, and the paternally expressed non-coding RNA gene KCNQ1OT1. KCNQ1OT1 and KCNQ1 are imprinted genes and are part of an imprinting control region (ICR). Mitsuya identified that KCNQ1OT1 is an antisense transcript of KCNQ1. KCNQ1OT1 is a paternally expressed allele and KCNQ1 is a maternally expressed allele. KCNQ1OT1 is a nuclear, 91 kb transcript, found in close proximity to the nucleolus in certain cell types.

Homeobox protein Nkx-2.2 is a protein that in humans is encoded by the NKX2-2 gene.
Natural antisense transcripts (NATs) are a group of RNAs encoded within a cell that have transcript complementarity to other RNA transcripts. They have been identified in multiple eukaryotes, including humans, mice, yeast and Arabidopsis thaliana. This class of RNAs includes both protein-coding and non-coding RNAs. Current evidence has suggested a variety of regulatory roles for NATs, such as RNA interference (RNAi), alternative splicing, genomic imprinting, and X-chromosome inactivation. NATs are broadly grouped into two categories based on whether they act in cis or in trans. Trans-NATs are transcribed from a different location than their targets and usually have complementarity to multiple transcripts with some mismatches. MicroRNAs (miRNA) are an example of trans-NATs that can target multiple transcripts with a few mismatches. Cis-natural antisense transcripts (cis-NATs) on the other hand are transcribed from the same genomic locus as their target but from the opposite DNA strand and form perfect pairs.

Long non-coding RNAs are a type of RNA, generally defined as transcripts more than 200 nucleotides that are not translated into protein. This arbitrary limit distinguishes long ncRNAs from small non-coding RNAs, such as microRNAs (miRNAs), small interfering RNAs (siRNAs), Piwi-interacting RNAs (piRNAs), small nucleolar RNAs (snoRNAs), and other short RNAs. Given that some lncRNAs have been reported to have the potential to encode small proteins or micro-peptides, the latest definition of lncRNA is a class of RNA molecules of over 200 nucleotides that have no or limited coding capacity. Long intervening/intergenic noncoding RNAs (lincRNAs) are sequences of lncRNA which do not overlap protein-coding genes.

HOTAIR is a human gene located between HOXC11 and HOXC12 on chromosome 12. It is the first example of an RNA expressed on one chromosome that has been found to influence transcription of HOXD cluster posterior genes located on chromosome 2. The sequence and function of HOTAIR is different in human and mouse. Sequence analysis of HOTAIR revealed that it exists in mammals, has poorly conserved sequences and considerably conserved structures, and has evolved faster than nearby HoxC genes. A subsequent study identified HOTAIR has 32 nucleotide long conserved noncoding element (CNE) that has a paralogous copy in HOXD cluster region, suggesting that the HOTAIR conserved sequences predates whole genome duplication events at the root of vertebrate. While the conserved sequence paralogous with HOXD cluster is 32 nucleotide long, the HOTAIR sequence conserved from human to fish is about 200 nucleotide long and is marked by active enhancer features.

Mediator complex subunit 13 is a protein that in humans is encoded by the MED13 gene.

MicroRNA 222 is a MicroRNA that in humans is encoded by the MIR222 gene, and is a known extracellular RNA (exRNA).
Epigenetics of human development is the study of how epigenetics effects human development.

MicroRNA 5008 is a protein that in humans is encoded by the MIR5008 gene.
MicroRNA 320b-2 is a protein that in humans is encoded by the MIR320B2 gene.

MicroRNA 6850 is a protein that in humans is encoded by the MIR6850 gene.

MicroRNA 203a is a small RNA that in humans is encoded by the preMIR203A gene.

MicroRNA 548f-4 is a protein that in humans is encoded by the MIR548F4 gene.

IFNG antisense RNA 1 is a long non-coding RNA that in humans is encoded by the IFNG-AS1 gene. It is a positive regulator of interferon gamma in T and NK cells.
References
- ↑ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ↑ "Entrez Gene: PTGS2 antisense NFKB1 complex-mediated expression regulator RNA" . Retrieved November 24, 2016.