A ribosomal protein is any of the proteins that, in conjunction with rRNA, make up the ribosomal subunits involved in the cellular process of translation. A large part of the knowledge about these organic molecules has come from the study of E. coli ribosomes. All ribosomal proteins have been isolated and many specific antibodies have been produced. These, together with electronic microscopy and the use of certain reactives, have allowed for the determination of the topography of the proteins in the ribosome. E. coli, other bacteria and Archaea have a 30S small subunit and a 50S large subunit, whereas humans and yeasts have a 40S small subunit and a 60S large subunit. Equivalent subunits are frequently numbered differently between bacteria, Archaea, yeasts and humans. More recently, a near-complete (near)atomic picture of the ribosomal proteins is emerging from the latest high-resolution cryo-EM data.

The Tryptophan operon leader is an RNA element found at the 5' of some bacterial tryptophan operons. The leader sequence can assume two different secondary structures known as the terminator and the anti-terminator structure. The leader also codes for very short peptide sequence that is rich in tryptophan. The terminator structure is recognised as a termination signal for RNA polymerase and the operon is not transcribed. This structure forms when the cell has an excess of tryptophan and ribosome movement over the leader transcript is not impeded. When there is a deficiency of the charged tryptophanyl tRNA the ribosome translating the leader peptide stalls and the antiterminator structure can form. This allows RNA polymerase to transcribe the operon.

The FMN riboswitch is a highly conserved RNA element that is found frequently in the 5'-untranslated regions of prokaryotic mRNAs that encode for flavin mononucleotide (FMN) biosynthesis and transport proteins. This element is a metabolite-dependent riboswitch that directly binds FMN in the absence of proteins. In Bacillus subtilis, the riboswitch controls gene expression by causing premature transcription termination within the 5' untranslated region of the ribDEAHT operon and precluding access to the ribosome-binding site of ypaA mRNA.

The bacterial glycine riboswitch is an RNA element that can bind the amino acid glycine. Glycine riboswitches usually consist of two metabolite-binding aptamer domains with similar structures in tandem. The aptamers were originally thought to cooperatively bind glycine to regulate the expression of downstream genes. In Bacillus subtilis, this riboswitch is found upstream of the gcvT operon which controls glycine degradation. It is thought that when glycine is in excess it will bind to both aptamers to activate these genes and facilitate glycine degradation.

The Lysine riboswitch is a metabolite binding RNA element found within certain messenger RNAs that serve as a precision sensor for the amino acid lysine. Allosteric rearrangement of mRNA structure is mediated by ligand binding, and this results in modulation of gene expression. Lysine riboswitch are most abundant in Firmicutes and Gammaproteobacteria where they are found upstream of a number of genes involved in lysine biosynthesis, transport and catabolism. The lysine riboswitch has also been identified independently and called the L box.
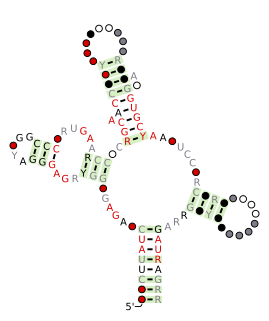
The SAM riboswitch is found upstream of a number of genes which code for proteins involved in methionine or cysteine biosynthesis in Gram-positive bacteria. Two SAM riboswitches in Bacillus subtilis that were experimentally studied act at the level of transcription termination control. The predicted secondary structure consists of a complex stem-loop region followed by a single stem-loop terminator region. An alternative and mutually exclusive form involves bases in the 3' segment of helix 1 with those in the 5' region of helix 5 to form a structure termed the anti-terminator form. When SAM is unbound, the anti-terminator sequence sequesters the terminator sequence so the terminator is unable to form, allowing the polymerase read-through the downstream gene. When the SAM is bound to the aptamer, the anti-terminator is sequestered by an anti-anti-terminator; the terminator forms and terminates the transcription. However, many SAM riboswitches are likely to regulate gene expression at the level of translation.
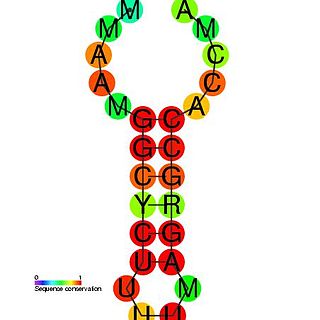
The histone 3' UTR stem-loop is an RNA element involved in nucleocytoplasmic transport of the histone mRNAs, and in the regulation of stability and of translation efficiency in the cytoplasm. The mRNAs of metazoan histone genes lack polyadenylation and a poly-A tail, instead 3' end processing occurs at a site between this highly conserved stem-loop and a purine rich region around 20 nucleotides downstream. The stem-loop is bound by a 31 kDa stem-loop binding protein. Together with U7 snRNA binding of the HDE, SLBP binding nucleates the formation of the processing complex.

Usually found in gram-positive bacteria, the T box leader sequence is an RNA element that controls gene expression through the regulation of translation by binding directly to a specific tRNA and sensing its aminoacylation state. This interaction controls expression of downstream aminoacyl-tRNA synthetase genes, amino acid biosynthesis, and uptake-related genes in a negative feedback loop. The uncharged tRNA acts as the effector for transcription antitermination of genes in the T-box leader family. The anticodon of a specific tRNA base pairs to a specifier sequence within the T-box motif, and the NCCA acceptor tail of the tRNA base pairs to a conserved bulge in the T-box antiterminator hairpin.
HutP is one of the anti-terminator proteins of Bacillus subtilis, which is responsible for regulating the expression of the hut structural genes of this organism in response to changes in the intracellular levels of L-histidine and divalent metal ions. In the hut operon, HutP is located just downstream from the promoter, while the five other subsequent structural genes, hutH, hutU, hutI, hutG and hutM, are positioned far downstream from the promoter. In the presence of L-histidine and divalent metal ions, HutP binds to the nascent hut mRNA leader transcript. This allows the anti-terminator to form, thereby preventing the formation of the terminator and permitting transcriptional read-through into the hut structural genes. In the absence of L-histidine and divalent metal ions or both, HutP does not bind to the hut mRNA, thus allowing the formation of a stem loop terminator structure within the nucleotide sequence located between the hutP and structural genes.
In a screen of the Bacillus subtilis genome for genes encoding ncRNAs, Saito et al. focused on 123 intergenic regions (IGRs) over 500 base pairs in length, the authors analyzed expression from these regions. Seven IGRs termed bsrC, bsrD, bsrE, bsrF, bsrG, bsrH and bsrI expressed RNAs smaller than 380 nt. All the small RNAs except BsrD RNA were expressed in transformed Escherichia coli cells harboring a plasmid with PCR-amplified IGRs of B. subtilis, indicating that their own promoters independently express small RNAs. Under non-stressed condition, depletion of the genes for the small RNAs did not affect growth. Although their functions are unknown, gene expression profiles at several time points showed that most of the genes except for bsrD were expressed during the vegetative phase, but undetectable during the stationary phase. Mapping the 5' ends of the 6 small RNAs revealed that the genes for BsrE, BsrF, BsrG, BsrH, and BsrI RNAs are preceded by a recognition site for RNA polymerase sigma factor σA.

The Bacillus-plasmid RNA motif is a predicted conserved RNA structure usually located in plasmids. It is known in species under the genera Bacillus and Lactobacillus. In Bacillus subtilis, it is found upstream of the hypothetical gene ydcS, whose function is unknown.

The pan RNA motif defines a conserved RNA structure that was identified using bioinformatics. pan motif RNAs are present in three phyla: Chloroflexi, Firmicutes and Proteobacteria, although within the latter phylum they are only known in deltaproteobacteria. A pan RNA is present in the Firmicute Bacillus subtilis, which is one of the most extensively studied bacteria.

The yjdF RNA motif is a conserved RNA structure identified using bioinformatics. Most yjdF RNAs are located in bacteria classified within the phylum Firmicutes. A yjdF RNA is found in the presumed 5' untranslated region of the yjdF gene in Bacillus subtilis, and almost all yjdF RNAs are found in the 5' UTRs of homologs of this gene. The function of the yjdF gene is unknown, but the protein that it is predicted to encode is classified by the Pfam Database as DUF2992.

The eps-Associated RNA element is a conserved RNA motif associated with exopolysaccharide (eps) or capsule biosynthesis genes in a subset of bacteria classified within the order Bacillales. It was initially discovered in Bacillus subtilis, located between the second and third gene in the eps operon. Deletion of the EAR element impairs biofilm formation.
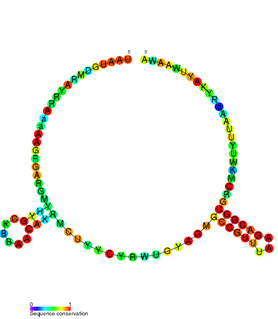
The TxpA/RatA toxin-antitoxin system was first identified in Bacillus subtilis. It consists of a non-coding 222nt sRNA called RatA and a protein toxin named TxpA.
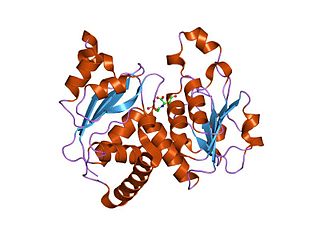
In molecular biology, the ATCase/OTCase family is a protein family which contains two related enzymes: aspartate carbamoyltransferase EC 2.1.3.2 and ornithine carbamoyltransferase EC 2.1.3.3. It has been shown that these enzymes are evolutionary related. The predicted secondary structure of both enzymes is similar and there are some regions of sequence similarities. One of these regions includes three residues which have been shown, by crystallographic studies to be implicated in binding the phosphoryl group of carbamoyl phosphate and may also play a role in trimerisation of the molecules. The N-terminal domain is the carbamoyl phosphate binding domain. The C-terminal domain is an aspartate/ornithine-binding domain.
In molecular biology, the PyrG leader is a cis-regulatory RNA element found at the 5' of the PyrG mRNA. The PyrG gene encodes a CTP synthase, which is involved in pyrimidine biosynthesis. The PyrG leader regulates expression of PyrG, PyrG can form into two different hairpin structures, a terminator or an anti-terminator. Under low CTP conditions, guanine (G) residues are incorporated at a specific site within the PyrG leader, these allow base-pairing with a uracil (U)-rich region and the formation of an anti-terminator loop, this results in increased expression of PyrG. Under high CTP conditions the guanines are not added, the anti-terminator loop cannot form and instead a terminator loop is formed, preventing further PyrG expression.
In molecular biology, the PyrC leader is a cis-regulatory RNA element found at the 5' of the PyrC mRNA in Enterobacteria. The PyrC gene encodes Dihydroorotase, an enzyme involved in pyrimidine biosynthesis. The PyrC leader regulates expression of PyrC. Translation initiation can occur at four different sites within this leader sequence, under high CTP conditions the translation initiation site is upstream of that used under low CTP conditions, additional cytosine residues are incorporated into the mRNA resulting in the formation of an RNA hairpin. This hairpin blocks ribosome-binding at the Shine-Dalgarno sequence, and therefore blocks expression of PyrC. Under low CTP conditions the initiation site is further downstream and does not result in hairpin formation, so the ribosome can bind to the Shine-Dalgarno sequence and PyrC is expressed.
In molecular biology, the PyrD leader is a cis-regulatory RNA element found at the 5' of the PyrC mRNA in Proteobacteria. The PyrD gene encodes dihydroorotate dehydrogenase, an enzyme involved in pyrimidine biosynthesis. The PyrD leader regulates expression of PyrD. Translation initiation can occur at more than one different site within this leader sequence, under high CTP or GTP conditions the translation initiation site is upstream of that used under low CTP/GTP conditions, additional cytosine residues are incorporated into the mRNA resulting in the formation of an RNA hairpin. This hairpin blocks ribosome-binding at the Shine-Dalgarno sequence, and therefore blocks expression of PyrD. Under low CTP/GTP conditions the initiation site is further downstream and does not result in hapirpin formation, so the ribosome can bind to the Shine-Dalgarno sequence and PyrD is expressed.
LysC is a prokaryotic aspartokinase involved in the biosynthesis of the amino acid lysine. It is found in a variety of bacteria, including Bacillus subtilis, Escherichia coli and Corynebacterium glutamicum. It is notable for containing a riboswitch, a structure in its messenger RNA that prevents its translation when bound to lysine. Such lysine riboswitch thus acts as a mechanism of negative feedback.















