
Antisense RNA (asRNA), also referred to as antisense transcript, natural antisense transcript (NAT) or antisense oligonucleotide, is a single stranded RNA that is complementary to a protein coding messenger RNA (mRNA) with which it hybridizes, and thereby blocks its translation into protein. asRNAs have been found in both prokaryotes and eukaryotes, antisense transcripts can be classified into short and long non-coding RNAs (ncRNAs). The primary function of asRNA is regulating gene expression. asRNAs may also be produced synthetically and have found wide spread use as research tools for gene knockdown. They may also have therapeutic applications.
In molecular biology and genetics, the sense of nucleic acid molecules is the nature of their roles and their complementary molecules' nucleic acid units' roles in specifying amino acids. Depending on the context within molecular biology, sense may have slightly different meanings.
The fertility factor allows genes to be transferred from one bacterium carrying the factor to another bacterium lacking the factor by conjugation. The F factor is carried on the F episome, the first episome to be discovered. Unlike other plasmids, F factor is constitutive for transfer proteins due to a mutation in the gene finO. The F plasmid belongs to a class of conjugative plasmids that control sexual functions of bacteria with a fertility inhibition (Fin) system.
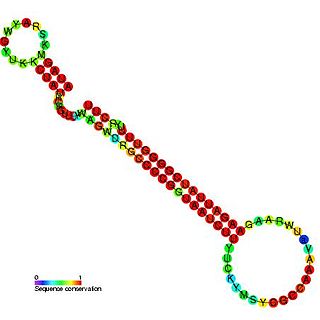
CopA-like RNA is a family of non-coding RNAs found on the R1 plasmid.
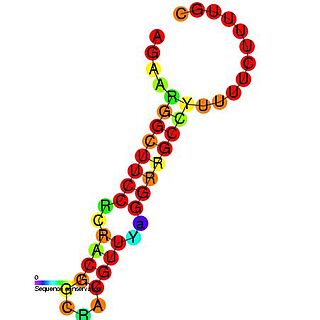
In molecular biology ctRNA is a plasmid encoded noncoding RNA that binds to the mRNA of repB and causes translational inhibition. ctRNA is encoded by plasmids and functions in rolling circle replication to maintain a low copy number. In Corynebacterium glutamicum, it achieves this by antisense pairing with the mRNA of RepB, a replication initiation protein. In Enterococcus faecium the plasmid pJB01 contains three open reading frames, copA, repB, and repC. The pJB01 ctRNA is coded on the opposite strand from the copA/repB intergenic region and partially overlaps an atypical ribosome binding site for repB.

FinP encodes an antisense non-coding RNA gene that is complementary to part of the TraJ 5' UTR. The FinOP system regulates the transfer of F-like plasmids. The traJ gene encodes a protein required for transcription from the major transfer promoter, pY. The FinO protein is essential for effective repression, acting by binding to FinP and protecting it from RNase E degradation.
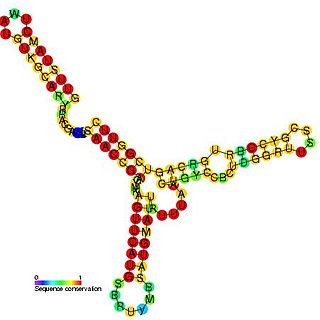
Anti-Q RNA is a small ncRNA from the conjugal plasmid pCF10 of Enterococcus faecalis. It is coded in cis to its regulatory target, prgQ, but can also act in trans. Anti-Q is known to interact with nascent prgQ transcripts to allow formation of an intrinsic terminator, or attenuator, thus preventing transcription of downstream genes. This mode of regulation is essentially the same as that of the countertranscript-driven attenuators that control copy number in pT181, pAMbeta1 and pIP501 and related Staphylococcal plasmids.
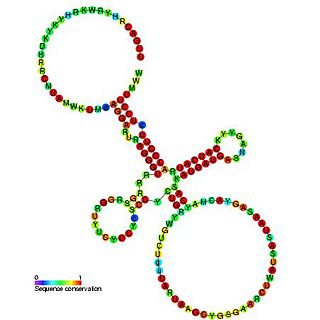
Sib RNA refers to a group of related non-coding RNA. They were originally named QUAD RNA after they were discovered as four repeat elements in Escherichia coli intergenic regions. The family was later renamed Sib when it was discovered that the number of repeats is variable in other species and in other E. coli strains.

The traJ 5' UTR is a cis acting RNA element which is involved in regulating plasmid transfer in bacteria.
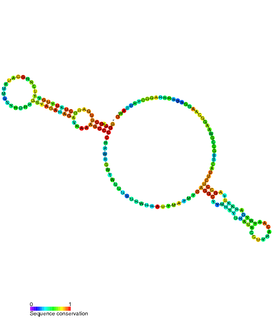
The hok/sok system is a postsegregational killing mechanism employed by the R1 plasmid in Escherichia coli. It was the first type I toxin-antitoxin pair to be identified through characterisation of a plasmid-stabilising locus. It is a type I system because the toxin is neutralised by a complementary RNA, rather than a partnered protein.
Iterons are directly repeated DNA sequences which play an important role in regulation of plasmid copy number in bacterial cells. It is one among the three negative regulatory elements found in plasmids which control its copy number. The others include antisense RNAs and ctRNAs. Iterons complex with cognate replication (Rep) initiator proteins to achieve the required regulatory effect.
Long non-coding RNAs are a type of RNA, defined as being transcripts with lengths exceeding 200 nucleotides that are not translated into protein. This somewhat arbitrary limit distinguishes long ncRNAs from small non-coding RNAs such as microRNAs (miRNAs), small interfering RNAs (siRNAs), Piwi-interacting RNAs (piRNAs), small nucleolar RNAs (snoRNAs), and other short RNAs. Long intervening/intergenic noncoding RNAs (lincRNAs) are sequences of lncRNA which do not overlap protein-coding genes.

The Bacillus-plasmid RNA motif is a predicted conserved RNA structure usually located in plasmids. It is known in species under the genera Bacillus and Lactobacillus. In Bacillus subtilis, it is found upstream of the hypothetical gene ydcS, whose function is unknown.

The lactis-plasmid RNA motif is a conserved RNA structure identified by bioinformatics. The RNAs are restricted to lactic acid bacteria, and are especially common in Lactococcus lactis. They typically lie near to repB genes, and are almost found in plasmids. This data suggested that lactis-plasmid RNAs participate in the control of plasmid abundance. However, many of the plasmids that carry lactis-plasmid RNAs also carry ctRNA-pND324 RNAs, which are involved in plasmid copy count regulation. Therefore lactis-plasmid RNAs might have a different function.
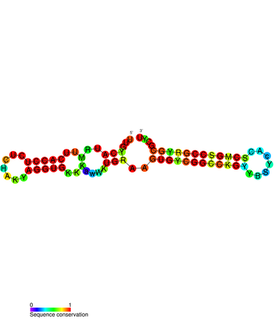
PtaRNA1 is a family of non-coding RNAs. Homologs of PtaRNA1 can be found in the proteobacteria families, Betaproteobacteria and Gammaproteobacteria. In all cases the PtaRNA1 is located anti-sense to a short protein-coding gene. In Xanthomonas campestris pv. vesicatoria, this gene is annotated as XCV2162 and is included in the plasmid toxin family of proteins.

A toxin-antitoxin system is a set of two or more closely linked genes that together encode both a "toxin" protein and a corresponding "antitoxin". When these systems are contained on plasmids – transferable genetic elements – they ensure that only the daughter cells that inherit the plasmid survive after cell division. If the plasmid is absent in a daughter cell, the unstable antitoxin is degraded and the stable toxic protein kills the new cell; this is known as 'post-segregational killing' (PSK). Toxin-antitoxin systems are widely distributed in prokaryotes, and organisms often have them in multiple copies.
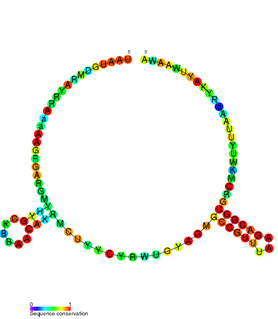
The TxpA/RatA toxin-antitoxin system was first identified in Bacillus subtilis. It consists of a non-coding 222nt sRNA called RatA and a protein toxin named TxpA.

The par stability determinant is a 400 bp locus of the pAD1 plasmid which encodes a type I toxin-antitoxin system in Enterococcus faecalis. It was the first such plasmid addiction module to be found in gram-positive bacteria.

miR-27 is a family of microRNA precursors found in animals, including humans. MicroRNAs are typically transcribed as ~70 nucleotide precursors and subsequently processed by the Dicer enzyme to give a ~22 nucleotide product. The excised region or, mature product, of the miR-27 precursor is the microRNA mir-27.
Plasmids must regulate their copy number to ensure that they do not excessively burden the host or become lost during cell division. Plasmids may be either high copy number plasmids or low copy number plasmids; the regulation mechanisms between these two types are often significantly different. Biotechnology applications may involve engineering plasmids to allow a very high copy number. For example, pBR322 is a low copy number plasmid from which several very high copy number cloning vectors have been derived.



















