Related Research Articles

Ribosomes ( ) are macromolecular machines, found within all cells, that perform biological protein synthesis. Ribosomes link amino acids together in the order specified by the codons of messenger RNA (mRNA) molecules to form polypeptide chains. Ribosomes consist of two major components: the small and large ribosomal subunits. Each subunit consists of one or more ribosomal RNA (rRNA) molecules and many ribosomal proteins. The ribosomes and associated molecules are also known as the translational apparatus.

Ribonuclease is a type of nuclease that catalyzes the degradation of RNA into smaller components. Ribonucleases can be divided into endoribonucleases and exoribonucleases, and comprise several sub-classes within the EC 2.7 and 3.1 classes of enzymes.

Ribonuclease H is a family of non-sequence-specific endonuclease enzymes that catalyze the cleavage of RNA in an RNA/DNA substrate via a hydrolytic mechanism. Members of the RNase H family can be found in nearly all organisms, from bacteria to archaea to eukaryotes.

Exonucleases are enzymes that work by cleaving nucleotides one at a time from the end (exo) of a polynucleotide chain. A hydrolyzing reaction that breaks phosphodiester bonds at either the 3′ or the 5′ end occurs. Its close relative is the endonuclease, which cleaves phosphodiester bonds in the middle (endo) of a polynucleotide chain. Eukaryotes and prokaryotes have three types of exonucleases involved in the normal turnover of mRNA: 5′ to 3′ exonuclease (Xrn1), which is a dependent decapping protein; 3′ to 5′ exonuclease, an independent protein; and poly(A)-specific 3′ to 5′ exonuclease.
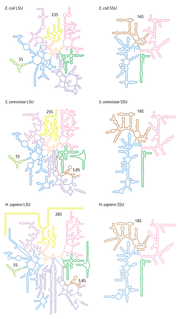
Ribosomal ribonucleic acid (rRNA) is a type of non-coding RNA which is the primary component of ribosomes, essential to all cells. rRNA is a ribozyme which carries out protein synthesis in ribosomes. Ribosomal RNA is transcribed from ribosomal DNA (rDNA) and then bound to ribosomal proteins to form small and large ribosome subunits. rRNA is the physical and mechanical factor of the ribosome that forces transfer RNA (tRNA) and messenger RNA (mRNA) to process and translate the latter into proteins. Ribosomal RNA is the predominant form of RNA found in most cells; it makes up about 80% of cellular RNA despite never being translated into proteins itself. Ribosomes are composed of approximately 60% rRNA and 40% ribosomal proteins by mass.

Transfer-messenger RNA is a bacterial RNA molecule with dual tRNA-like and messenger RNA-like properties. The tmRNA forms a ribonucleoprotein complex (tmRNP) together with Small Protein B (SmpB), Elongation Factor Tu (EF-Tu), and ribosomal protein S1. In trans-translation, tmRNA and its associated proteins bind to bacterial ribosomes which have stalled in the middle of protein biosynthesis, for example when reaching the end of a messenger RNA which has lost its stop codon. The tmRNA is remarkably versatile: it recycles the stalled ribosome, adds a proteolysis-inducing tag to the unfinished polypeptide, and facilitates the degradation of the aberrant messenger RNA. In the majority of bacteria these functions are carried out by standard one-piece tmRNAs. In other bacterial species, a permuted ssrA gene produces a two-piece tmRNA in which two separate RNA chains are joined by base-pairing.
Bacterial translation is the process by which messenger RNA is translated into proteins in bacteria.
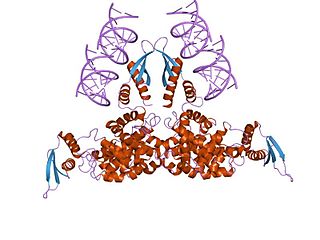
Ribonuclease III (BRENDA 3.1.26.3) is a type of ribonuclease that recognizes dsRNA and cleaves it at specific targeted locations to transform them into mature RNAs. These enzymes are a group of endoribonucleases that are characterized by their ribonuclease domain, which is labelled the RNase III domain. They are ubiquitous compounds in the cell and play a major role in pathways such as RNA precursor synthesis, RNA Silencing, and the pnp autoregulatory mechanism.
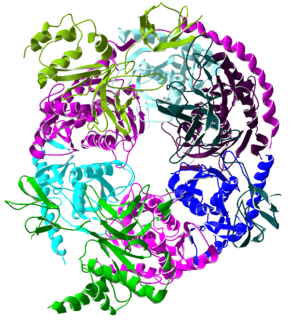
The exosome complex is a multi-protein intracellular complex capable of degrading various types of RNA molecules. Exosome complexes are found in both eukaryotic cells and archaea, while in bacteria a simpler complex called the degradosome carries out similar functions.

The 5S ribosomal RNA is an approximately 120 nucleotide-long ribosomal RNA molecule with a mass of 40 kDa. It is a structural and functional component of the large subunit of the ribosome in all domains of life, with the exception of mitochondrial ribosomes of fungi and animals. The designation 5S refers to the molecule's sedimentation velocity in an ultracentrifuge, which is measured in Svedberg units (S).
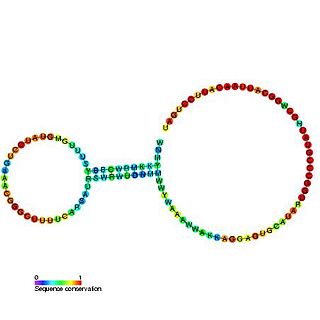
The alpha operon ribosome binding site in bacteria is surrounded by this complex pseudoknotted RNA structure. Translation of the mRNA produces 4 ribosomal protein products, one of which (S4) acts as a translational repressor by binding to the nested pseudoknot region. The mechanism of repression is thought to involve a conformational switch in the pseudoknot region and ribosome entrapment.

An exoribonuclease is an exonuclease ribonuclease, which are enzymes that degrade RNA by removing terminal nucleotides from either the 5' end or the 3' end of the RNA molecule. Enzymes that remove nucleotides from the 5' end are called 5'-3' exoribonucleases, and enzymes that remove nucleotides from the 3' end are called 3'-5' exoribonucleases.

Non-stop decay (NSD) is a cellular mechanism of mRNA surveillance to detect mRNA molecules lacking a stop codon and prevent these mRNAs from translation. The non-stop decay pathway releases ribosomes that have reached the far 3' end of an mRNA and guides the mRNA to the exosome complex, or to RNase R in bacteria for selective degradation. In contrast to Nonsense-mediated decay (NMD), polypeptides do not release from the ribosome, and thus, NSD seems to involve mRNA decay factors distinct from NMD.

(p)ppGpp, guanosine pentaphosphate and tetraphosphate, also known as the "magic spot" nucleotides, are alarmones involved in the stringent response in bacteria that cause the inhibition of RNA synthesis when there is a shortage of amino acids. This inhibition by (p)ppGpp decreases translation in the cell, conserving amino acids present. Furthermore, ppGpp and pppGpp cause the up-regulation of many other genes involved in stress response such as the genes for amino acid uptake and biosynthesis.
The degradosome is a multiprotein complex present in most bacteria that is involved in the processing of ribosomal RNA and the degradation of messenger RNA and is regulated by Non-coding RNA. It contains the proteins RNA helicase B, RNase E and Polynucleotide phosphorylase.
Ribosomal frameshifting, also known as translational frameshifting or translational recoding, is a biological phenomenon that occurs during translation that results in the production of multiple, unique proteins from a single mRNA. The process can be programmed by the nucleotide sequence of the mRNA and is sometimes affected by the secondary, 3-dimensional mRNA structure. It has been described mainly in viruses, retrotransposons and bacterial insertion elements, and also in some cellular genes.
EF-Ts is one of the prokaryotic elongation factors. It is found in human mitochrondria as TSFM. It is similar to eukaryotic EF-1B.

ATP-binding cassette sub-family E member 1 (ABCE1) also known as RNase L inhibitor (RLI) is an enzyme that in humans is encoded by the ABCE1 gene.
Ribonuclease E is a bacterial ribonuclease that participates in the processing of ribosomal RNA and the chemical degradation of bulk cellular RNA.

Ribonuclease T is a ribonuclease enzyme involved in the maturation of transfer RNA and ribosomal RNA in bacteria, as well as in DNA repair pathways. It is a member of the DnaQ family of exonucleases and non-processively acts on the 3' end of single-stranded nucleic acids. RNase T is capable of cleaving both DNA and RNA, with extreme sequence specificity discriminating against cytosine at the 3' end of the substrate.
References
- ↑ Cheng ZF, Deutscher MP (January 2005). "An important role for RNase R in mRNA decay". Molecular Cell. 17 (2): 313–8. doi: 10.1016/j.molcel.2004.11.048 . PMID 15664199.
- ↑ Venkataraman K, Guja KE, Garcia-Diaz M, Karzai AW (2014). "Non-stop mRNA decay: a special attribute of trans-translation mediated ribosome rescue". Frontiers in Microbiology. 5: 93. doi: 10.3389/fmicb.2014.00093 . PMC 3949413 . PMID 24653719.
- 1 2 3 Domingues S, Moreira RN, Andrade JM, Dos Santos RF, Bárria C, Viegas SC, Arraiano CM (July 2015). "The role of RNase R in trans-translation and ribosomal quality control". Biochimie. 114: 113–8. doi:10.1016/j.biochi.2014.12.012. PMID 25542646.
- 1 2 Suzuki H, Tsukahara T (May 2014). "A view of pre-mRNA splicing from RNase R resistant RNAs". International Journal of Molecular Sciences. 15 (6): 9331–42. doi: 10.3390/ijms15069331 . PMC 4100097 . PMID 24865493.
- ↑ Cheng ZF, Deutscher MP (June 2002). "Purification and characterization of the Escherichia coli exoribonuclease RNase R. Comparison with RNase II". The Journal of Biological Chemistry. 277 (24): 21624–9. doi: 10.1074/jbc.M202942200 . PMID 11948193.
- ↑ Vincent HA, Deutscher MP (October 2006). "Substrate recognition and catalysis by the exoribonuclease RNase R". The Journal of Biological Chemistry. 281 (40): 29769–75. doi: 10.1074/jbc.M606744200 . PMID 16893880.