GTPases are a large family of hydrolase enzymes that bind to the nucleotide guanosine triphosphate (GTP) and hydrolyze it to guanosine diphosphate (GDP). The GTP binding and hydrolysis takes place in the highly conserved P-loop "G domain", a protein domain common to many GTPases.
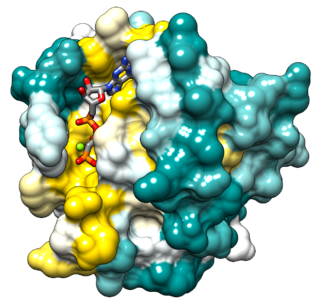
Ras, from "Rat sarcoma virus", is a family of related proteins that are expressed in all animal cell lineages and organs. All Ras protein family members belong to a class of protein called small GTPase, and are involved in transmitting signals within cells. Ras is the prototypical member of the Ras superfamily of proteins, which are all related in three-dimensional structure and regulate diverse cell behaviours.

Guanosine diphosphate, abbreviated GDP, is a nucleoside diphosphate. It is an ester of pyrophosphoric acid with the nucleoside guanosine. GDP consists of a pyrophosphate group, a pentose sugar ribose, and the nucleobase guanine.
GTPase-activating proteins or GTPase-accelerating proteins (GAPs) are a family of regulatory proteins whose members can bind to activated G proteins and stimulate their GTPase activity, with the result of terminating the signaling event. GAPs are also known as RGS protein, or RGS proteins, and these proteins are crucial in controlling the activity of G proteins. Regulation of G proteins is important because these proteins are involved in a variety of important cellular processes. The large G proteins, for example, are involved in transduction of signaling from the G protein-coupled receptor for a variety of signaling processes like hormonal signaling, and small G proteins are involved in processes like cellular trafficking and cell cycling. GAP's role in this function is to turn the G protein's activity off. In this sense, GAPs function is opposite to that of guanine nucleotide exchange factors (GEFs), which serve to enhance G protein signaling.

ADP ribosylation factors (ARFs) are members of the ARF family of GTP-binding proteins of the Ras superfamily. ARF family proteins are ubiquitous in eukaryotic cells, and six highly conserved members of the family have been identified in mammalian cells. Although ARFs are soluble, they generally associate with membranes because of N-terminus myristoylation. They function as regulators of vesicular traffic and actin remodelling.
Phospholipase D (EC 3.1.4.4, lipophosphodiesterase II, lecithinase D, choline phosphatase, PLD; systematic name phosphatidylcholine phosphatidohydrolase) is an enzyme of the phospholipase superfamily that catalyses the following reaction

EF-Tu is a prokaryotic elongation factor responsible for catalyzing the binding of an aminoacyl-tRNA (aa-tRNA) to the ribosome. It is a G-protein, and facilitates the selection and binding of an aa-tRNA to the A-site of the ribosome. As a reflection of its crucial role in translation, EF-Tu is one of the most abundant and highly conserved proteins in prokaryotes. It is found in eukaryotic mitochondria as TUFM.

Guanine nucleotide exchange factors (GEFs) are proteins or protein domains that activate monomeric GTPases by stimulating the release of guanosine diphosphate (GDP) to allow binding of guanosine triphosphate (GTP). A variety of unrelated structural domains have been shown to exhibit guanine nucleotide exchange activity. Some GEFs can activate multiple GTPases while others are specific to a single GTPase.
Elongation factor 4 (EF-4) is an elongation factor that is thought to back-translocate on the ribosome during the translation of RNA to proteins. It is found near-universally in bacteria and in eukaryotic endosymbiotic organelles including the mitochondria and the plastid. Responsible for proofreading during protein synthesis, EF-4 is a recent addition to the nomenclature of bacterial elongation factors.
The Rho family of GTPases is a family of small signaling G proteins, and is a subfamily of the Ras superfamily. The members of the Rho GTPase family have been shown to regulate many aspects of intracellular actin dynamics, and are found in all eukaryotic kingdoms, including yeasts and some plants. Three members of the family have been studied in detail: Cdc42, Rac1, and RhoA. All G proteins are "molecular switches", and Rho proteins play a role in organelle development, cytoskeletal dynamics, cell movement, and other common cellular functions.

Cell division control protein 42 homolog is a protein that in humans is encoded by the CDC42 gene. Cdc42 is involved in regulation of the cell cycle. It was originally identified in S. cerevisiae (yeast) as a mediator of cell division, and is now known to influence a variety of signaling events and cellular processes in a variety of organisms from yeast to mammals.

Adenylylation, more commonly known as AMPylation, is a process in which an adenosine monophosphate (AMP) molecule is covalently attached to the amino acid side chain of a protein. This covalent addition of AMP to a hydroxyl side chain of the protein is a post-translational modification. Adenylylation involves a phosphodiester bond between a hydroxyl group of the molecule undergoing adenylylation, and the phosphate group of the adenosine monophosphate nucleotide. Enzymes that are capable of catalyzing this process are called AMPylators.

Transforming protein RhoA, also known as Ras homolog family member A (RhoA), is a small GTPase protein in the Rho family of GTPases that in humans is encoded by the RHOA gene. While the effects of RhoA activity are not all well known, it is primarily associated with cytoskeleton regulation, mostly actin stress fibers formation and actomyosin contractility. It acts upon several effectors. Among them, ROCK1 and DIAPH1 are the best described. RhoA, and the other Rho GTPases, are part of a larger family of related proteins known as the Ras superfamily, a family of proteins involved in the regulation and timing of cell division. RhoA is one of the oldest Rho GTPases, with homologues present in the genomes since 1.5 billion years. As a consequence, RhoA is somehow involved in many cellular processes which emerged throughout evolution. RhoA specifically is regarded as a prominent regulatory factor in other functions such as the regulation of cytoskeletal dynamics, transcription, cell cycle progression and cell transformation.
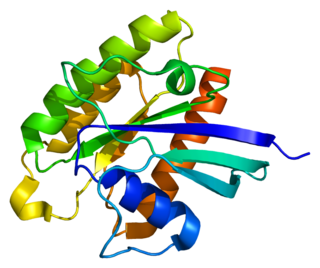
RHEB also known as Ras homolog enriched in brain (RHEB) is a GTP-binding protein that is ubiquitously expressed in humans and other mammals. The protein is largely involved in the mTOR pathway and the regulation of the cell cycle.

EF-G is a prokaryotic elongation factor involved in protein translation. As a GTPase, EF-G catalyzes the movement (translocation) of transfer RNA (tRNA) and messenger RNA (mRNA) through the ribosome.

Dishevelled (Dsh) is a family of proteins involved in canonical and non-canonical Wnt signalling pathways. Dsh is a cytoplasmic phosphoprotein that acts directly downstream of frizzled receptors. It takes its name from its initial discovery in flies, where a mutation in the dishevelled gene was observed to cause improper orientation of body and wing hairs. There are vertebrate homologs in zebrafish, Xenopus (Xdsh), mice and humans. Dsh relays complex Wnt signals in tissues and cells, in normal and abnormal contexts. It is thought to interact with the SPATS1 protein when regulating the Wnt Signalling pathway.
The Walker A and Walker B motifs are protein sequence motifs, known to have highly conserved three-dimensional structures. These were first reported in ATP-binding proteins by Walker and co-workers in 1982.
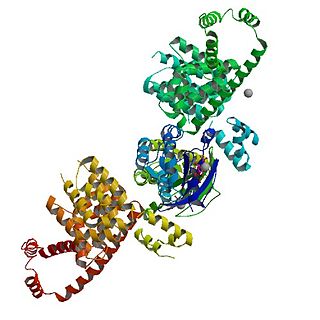
mDia1 is a member of the protein family called the formins and is a Rho effector. It is the mouse version of the diaphanous homolog 1 of Drosophila. mDia1 localizes to cells' mitotic spindle and midbody, plays a role in stress fiber and filopodia formation, phagocytosis, activation of serum response factor, formation of adherens junctions, and it can act as a transcription factor. mDia1 accelerates actin nucleation and elongation by interacting with barbed ends of actin filaments. The gene encoding mDia1 is located on Chromosome 18 of Mus musculus and named Diap1.

Ras and EF-hand domain-containing protein also known as Ras-related protein Rab-45 is a protein that in humans is encoded by the RASEF gene.
Alfred "Fred" Wittinghofer is a German biochemist.













