
Sinorhizobium meliloti is a Gram-negative bacterium which fixes atmospheric nitrogen. It forms a symbiotic relationship with legumes from the genera Medicago, Melilotus and Trigonella, including the model legume Medicago truncatula. This symbiosis results in a new plant organ termed a root nodule and is deemed symbiotic as it leaves excess nitrogen behind for the plant. S. meliloti are mobile and possess a cluster of peritrichous flagella. The S. meliloti genome contains four genes coding for flagellin. These include fliC1C2–fliC3C4. The genome contains three replicons: a chromosome, a chromid, and a plasmid. Individual strains may possess additional, accessory plasmids. Five S. meliloti genomes have been sequenced to date: Rm1021, AK83, BL225C, Rm41, and SM11 with 1021 considered to be the wild type.
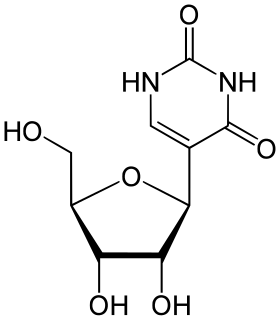
Pseudouridine is an isomer of the nucleoside uridine in which the uracil is attached via a carbon-carbon instead of a nitrogen-carbon glycosidic bond. Pseudouridine is the most abundant RNA modification in cellular RNA. After transcription and following synthesis, RNA can be modified with over 100 chemically distinct modifications. These can potentially regulate RNA expression post-transcriptionally, in addition to the four standard nucleotides and play a variety of roles in the cell including translation, localization and stabilization of RNA. Pseudouridine, being one of them, is the C5-glycoside isomer of uridine that contains a C-C bond between C1 of the ribose sugar and C5 of uracil, rather than usual C1-N1 bond found in uridine. The C-C bond gives it more rotational freedom and conformational flexibility. In addition, pseudouridine has an extra hydrogen bond donor at the N1 position. Also known as 5-ribosyluracil, pseudouridine is a ubiquitous yet enigmatic constituent of structural RNAs. Recently it has also been discovered in coding RNA. Being the most abundant, it is found in all 3 phylogenetic domains of life and was the first to be discovered. This nucleotide is considered to be the “fifth nucleotide”. It accounts for 4% of the nucleotides in the yeast tRNA. This base modification is able to stabilize RNA and improve base-stacking by forming additional hydrogen bonds with water through its extra imino group. There are 11 pseudouridines in the Escherichia coli rRNA, 30 in yeast cytoplasmic rRNA and a single modification in mitochondrial 21S rRNA and about 100 pseudouridines in human rRNA indicating that the extent of pseudouridylation increases with the complexity of an organism. Pseudouridine in rRNA and tRNA has been shown to fine-tune and stabilize the regional structure and help maintain their functions in mRNA decoding, ribosome assembly, processing and translation. Pseudouridine in snRNA has been shown to enhance spliceosomal RNA-pre-mRNA interaction to facilitate splicing regulation.

suhB later renamed mmgR was a putative non-coding RNA found multiple times in the Agrobacterium tumefaciens genome and related alpha-proteobacteria. Other non-coding RNAs uncovered in the same analysis include: speF, ybhL, metA, and serC.
Sinorhizobium/Ensifer is a genus of nitrogen-fixing bacteria (rhizobia), three of which have been sequenced.
Autoinducers are signaling molecules that are produced in response to changes in cell-population density. As the density of quorum sensing bacterial cells increases so does the concentration of the autoinducer. Detection of signal molecules by bacteria acts as stimulation which leads to altered gene expression once the minimal threshold is reached. Quorum sensing is a phenomenon that allows both Gram-negative and Gram-positive bacteria to sense one another and to regulate a wide variety of physiological activities. Such activities include symbiosis, virulence, motility, antibiotic production, and biofilm formation. Autoinducers come in a number of different forms depending on the species, but the effect that they have is similar in many cases. Autoinducers allow bacteria to communicate both within and between different species. This communication alters gene expression and allows bacteria to mount coordinated responses to their environments, in a manner that is comparable to behavior and signaling in higher organisms. Not surprisingly, it has been suggested that quorum sensing may have been an important evolutionary milestone that ultimately gave rise to multicellular life forms.
Bacterial small RNAs (sRNA) are small RNAs produced by bacteria; they are 50- to 500-nucleotide non-coding RNA molecules, highly structured and containing several stem-loops. Numerous sRNAs have been identified using both computational analysis and laboratory-based techniques such as Northern blotting, microarrays and RNA-Seq in a number of bacterial species including Escherichia coli, the model pathogen Salmonella, the nitrogen-fixing alphaproteobacterium Sinorhizobium meliloti, marine cyanobacteria, Francisella tularensis, Streptococcus pyogenes, the pathogen Staphylococcus aureus, and the plant pathogen Xanthomonas oryzae pathovar oryzae. Bacterial sRNAs affect how genes are expressed within bacterial cells via interaction with mRNA or protein, and thus can affect a variety of bacterial functions like metabolism, virulence, environmental stress response, and structure.
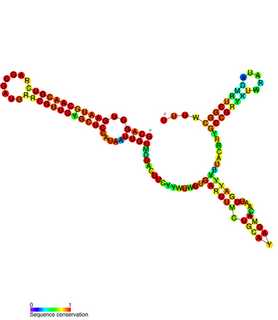
FnrS RNA is a family of Hfq-binding small RNA whose expression is upregulated in response to anaerobic conditions. It is named FnrS because its expression is strongly dependent on fumarate and nitrate reductase regulator (FNR), a direct oxygen availability sensor.
Rsa RNAs are non-coding RNAs found in the bacterium Staphylococcus aureus. The shared name comes from their discovery, and does not imply homology. Bioinformatics scans identified the 16 Rsa RNA families named RsaA-K and RsaOA-OG. Others, RsaOH-OX, were found thanks to an RNomic approach. Although the RNAs showed varying expression patterns, many of the newly discovered RNAs were shown to be Hfq-independent and most carried a C-rich motif (UCCC).

enod40, also known as early nodulin 40, is a gene found in flowering plants. The gene has characteristics of both protein and Non-coding RNA genes. There is some evidence that the non-coding characteristics of this gene are more widely conserved than the protein coding sequences. In soyabeans enod40 was found to be expressed during early stages of formation of nitrogen-fixing root nodules that are associated with symbiotic soil rhizobial bacteria. The gene is also active in roots containing fungi forming phosphate-acquiring arbuscular mycorrhiza. An interaction with a novel RNA-binding protein MtRBP1 investigated in the development of Root nodule suggests ENOD40 has a function of cytoplasmic relocalization of nuclear proteins. In the study of non-legume plants, the over-expression of ENOD40 in transgenic Arabidopsis lines was observed a reduction of cell expansion.
VapBC is the largest family of type II toxin-antitoxin system genetic loci in prokaryotes. VapBC operons consist of two genes: VapC encodes a toxic PilT N-terminus (PIN) domain, and VapB encodes a matching antitoxin. The toxins in this family are thought to perform RNA cleavage, which is inhibited by the co-expression of the antitoxin, in a manner analogous to a poison and antidote.

Post-genomic research has rendered bacterial small non-coding RNAs (sRNAs) as major players in post-transcriptional regulation of gene expression in response to environmental stimuli. The α-subdivision of the Proteobacteria includes Gram-negative microorganisms with diverse life styles; frequently involving long-term interactions with higher eukaryotes.
αr7 is a family of bacterial small non-coding RNAs with representatives in a broad group of α-proteobacterial species from the order Rhizobiales. The first member of this family was found in a Sinorhizobium meliloti 1021 locus located in the chromosome (C). Further homology and structure conservation analysis identified full-length homologs in several nitrogen-fixing symbiotic rhizobia, in the plant pathogens belonging to Agrobacterium species as well as in a broad spectrum of Brucella species. αr7 RNA species are 134-159 nucleotides (nt) long and share a well defined common secondary structure. αr7 transcripts can be catalogued as trans-acting sRNAs expressed from well-defined promoter regions of independent transcription units within intergenic regions (IGRs) of the α-proteobacterial genomes.
αr9 is a family of bacterial small non-coding RNAs with representatives in a broad group of α-proteobacteria from the order Rhizobiales. The first member of this family (Smr9C) was found in a Sinorhizobium meliloti 1021 locus located in the chromosome (C). Further homology and structure conservation analysis have identified full-length Smr9C homologs in several nitrogen-fixing symbiotic rhizobia, in the plant pathogens belonging to Agrobacterium species as well as in a broad spectrum of Brucella species. αr9C RNA species are 144-158 nt long and share a well defined common secondary structure consisting of seven conserved regions. Most of the αr9 transcripts can be catalogued as trans-acting sRNAs expressed from well-defined promoter regions of independent transcription units within intergenic regions (IGRs) of the α-proteobacterial genomes.
αr14 is a family of bacterial small non-coding RNAs with representatives in a broad group of α-proteobacteria. The first member of this family (Smr14C2) was found in a Sinorhizobium meliloti 1021 locus located in the chromosome (C). It was later renamed NfeR1 and shown to be highly expressed in salt stress and during the symbiotic interaction on legume roots. Further homology and structure conservation analysis identified 2 other chromosomal copies and 3 plasmidic ones. Moreover, full-length Smr14C homologs have been identified in several nitrogen-fixing symbiotic rhizobia, in the plant pathogens belonging to Agrobacterium species as well as in a broad spectrum of Brucella species. αr14C RNA species are 115-125 nt long and share a well defined common secondary structure. Most of the αr14 transcripts can be catalogued as trans-acting sRNAs expressed from well-defined promoter regions of independent transcription units within intergenic regions (IGRs) of the α-proteobacterial genomes.
αr15 is a family of bacterial small non-coding RNAs with representatives in a broad group of α-proteobacteria from the order Rhizobiales. The first members of this family were found tandemly arranged in the same intergenic region (IGR) of the Sinorhizobium meliloti 1021 chromosome (C). Further homology and structure conservation analysis have identified full-length Smr15C1 and Smr15C2 homologs in several nitrogen-fixing symbiotic rhizobia, in the plant pathogens belonging to Agrobacterium species as well as in a broad spectrum of Brucella species. The Smr15C1 and Smr15C2 homologs are also encoded in tandem within the same IGR region of Rhizobium and Agrobacterium species, whereas in Brucella species the αr15C loci are spread in the IGRs of Chromosome I. Moreover, this analysis also identified a third αr15 loci in extrachromosomal replicons of the mentioned nitrogen-fixing α-proteobacteria and in the Chromosome II of Brucella species. αr15 RNA species are 99-121 nt long and share a well defined common secondary structure consisting of three stem loops. The transcripts of the αr15 family can be catalogued as trans-acting sRNAs encoded by independent transcription units with recognizable promoter and transcription termination signatures within intergenic regions (IGRs) of the α-proteobacterial genomes.
αr35 is a family of bacterial small non-coding RNAs with representatives in a reduced group of α-proteobacteria from the order Rhizobiales. The first member of this family (Smr35B) was found in a Sinorhizobium meliloti 1021 locus located in the symbiotic plasmid B (pSymB). Further homology and structure conservation analysis have identified full-length SmrB35 homologs in other legume symbionts, as well as in the human and plant pathogens Ochrobactrum anthropi and Agrobacterium tumefaciens, respectively. αr35 RNA species are 139-142 nt long and share a common secondary structure consisting of two stem loops and a well conserved rho independent terminator. Most of the αr35 transcripts can be catalogued as trans-acting sRNAs expressed from well-defined promoter regions of independent transcription units within intergenic regions of the α-proteobacterial genomes.
αr45 is a family of bacterial small non-coding RNAs with representatives in a broad group of α-proteobacteria from the order Rhizobiales. The first member of this family (Smr45C) was found in a Sinorhizobium meliloti 1021 locus located in the chromosome (C). Further homology and structure conservation analysis identified homologs in several nitrogen-fixing symbiotic rhizobia, in the plant pathogens belonging to Agrobacterium species as well as in a broad spectrum of Brucella species, in Bartonella species, in several members of the Xanthobactereacea family, and in some representatives of the Beijerinckiaceae family. αr45C RNA species are 147-153 nt long and share a well defined common secondary structure. All of the αr45 transcripts can be catalogued as trans-acting sRNAs expressed from well-defined promoter regions of independent transcription units within intergenic regions (IGRs) of the α-proteobacterial genomes.

4-Hydroxy-tetrahydrodipicolinate synthase (EC 4.3.3.7, dihydrodipicolinate synthase, dihydropicolinate synthetase, dihydrodipicolinic acid synthase, L-aspartate-4-semialdehyde hydro-lyase (adding pyruvate and cyclizing), dapA (gene)) is an enzyme with the systematic name L-aspartate-4-semialdehyde hydro-lyase (adding pyruvate and cyclizing; (4S)-4-hydroxy-2,3,4,5-tetrahydro-(2S)-dipicolinate-forming). This enzyme catalyses the following chemical reaction
EcpR1 is a trans-encoded small non-coding RNA in the plant-symbiotic Sinorhizobium meliloti, previously named SmelC291, SmrC10, or Sra33. According to its overproduction phenotype it was renamed Elongated Cell Phenotype RNA1. Induced by stress, EcpR1 negatively regulates cell cycle master regulatory genes dnaA and gcrA at post-transcriptional level by base pairing between its strongly conserved GC-rich loop and the target mRNAs. It is suggested that EcpR1 connects stress adaptation and cell cycle progression.

The chrB-a RNA motif and chrB-b RNA motif refer to a related, conserved RNA structure that was discovered by bioinformatics. The structures of these motifs are similar, and some genomic locations are predicted to exhibit both motifs. The chrB-b motif has an extra pseudoknot that is not consistently found in chrB-a examples. It was proposed that the two motifs could be unified into one common structure, with additional information.










