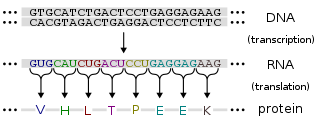
The genetic code is the set of rules used by living cells to translate information encoded within genetic material into proteins. Translation is accomplished by the ribosome, which links amino acids in an order specified by messenger RNA (mRNA), using transfer RNA (tRNA) molecules to carry amino acids and to read the mRNA three nucleotides at a time. The genetic code is highly similar among all organisms and can be expressed in a simple table with 64 entries.

Selenocysteine is the 21st proteinogenic amino acid.
Codon usage bias refers to differences in the frequency of occurrence of synonymous codons in coding DNA. A codon is a series of three nucleotides that encodes a specific amino acid residue in a polypeptide chain or for the termination of translation.
In genetics, an expressed sequence tag (EST) is a short sub-sequence of a cDNA sequence. ESTs may be used to identify gene transcripts, and are instrumental in gene discovery and in gene-sequence determination. The identification of ESTs has proceeded rapidly, with approximately 74.2 million ESTs now available in public databases.

In molecular biology, a reading frame is a way of dividing the sequence of nucleotides in a nucleic acid molecule into a set of consecutive, non-overlapping triplets. Where these triplets equate to amino acids or stop signals during translation, they are called codons.

A frameshift mutation is a genetic mutation caused by indels of a number of nucleotides in a DNA sequence that is not divisible by three. Due to the triplet nature of gene expression by codons, the insertion or deletion can change the reading frame, resulting in a completely different translation from the original. The earlier in the sequence the deletion or insertion occurs, the more altered the protein. A frameshift mutation is not the same as a single-nucleotide polymorphism in which a nucleotide is replaced, rather than inserted or deleted. A frameshift mutation will in general cause the reading of the codons after the mutation to code for different amino acids. The frameshift mutation will also alter the first stop codon encountered in the sequence. The polypeptide being created could be abnormally short or abnormally long, and will most likely not be functional.
The Crick, Brenner et al. experiment (1961) was a scientific experiment performed by Francis Crick, Sydney Brenner, Leslie Barnett and R.J. Watts-Tobin. This study demonstrated that the genetic code is made up of a series of three base pair codons which code for individual amino acids. The experiment also elucidated the nature of gene expression and frame-shift mutations.
This is a list of topics in molecular biology. See also index of biochemistry articles.

In genetics, an insertion is the addition of one or more nucleotide base pairs into a DNA sequence. This can often happen in microsatellite regions due to the DNA polymerase slipping. Insertions can be anywhere in size from one base pair incorrectly inserted into a DNA sequence to a section of one chromosome inserted into another. The mechanism of the smallest single base insertion mutations is believed to be through base-pair separation between the template and primer strands followed by non-neighbor base stacking, which can occur locally within the DNA polymerase active site. On a chromosome level, an insertion refers to the insertion of a larger sequence into a chromosome. This can happen due to unequal crossover during meiosis.
The start codon is the first codon of a messenger RNA (mRNA) transcript translated by a ribosome. The start codon always codes for methionine in eukaryotes and a modified Met (fMet) in prokaryotes. The most common start codon is AUG.
Eukaryotic translation is the biological process by which messenger RNA is translated into proteins in eukaryotes. It consists of four phases: initiation, elongation, termination, and recycling.
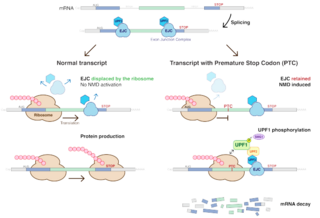
Nonsense-mediated mRNA decay (NMD) is a surveillance pathway that exists in all eukaryotes. Its main function is to reduce errors in gene expression by eliminating mRNA transcripts that contain premature stop codons. Translation of these aberrant mRNAs could, in some cases, lead to deleterious gain-of-function or dominant-negative activity of the resulting proteins.
Cis-regulatory elements (CREs) are regions of non-coding DNA which regulate the transcription of neighboring genes. CREs are vital components of genetic regulatory networks, which in turn control morphogenesis, the development of anatomy, and other aspects of embryonic development, studied in evolutionary developmental biology.
This glossary of genetics is a list of definitions of terms and concepts commonly used in the study of genetics and related disciplines in biology, including molecular biology and evolutionary biology. It is intended as introductory material for novices; for more specific and technical detail, see the article corresponding to each term.

HIV ribosomal frameshift signal is a ribosomal frameshift (PRF) that human immunodeficiency virus (HIV) uses to translate several different proteins from the same sequence.
Ribosomal frameshifting, also known as translational frameshifting or translational recoding, is a biological phenomenon that occurs during translation that results in the production of multiple, unique proteins from a single mRNA. The process can be programmed by the nucleotide sequence of the mRNA and is sometimes affected by the secondary, 3-dimensional mRNA structure. It has been described mainly in viruses, retrotransposons and bacterial insertion elements, and also in some cellular genes.
John Atkins is a research professor at University College Cork and a member of the Royal Irish Academy since 2003. Atkins was the first Irish national to be elected as a member of the EMBO Organization. In 2002 Science Foundation Ireland appointed Atkins as its first Director of Biotechnology. Atkins is also an honorary Professor of Genetics at his alma mater Trinity College, Dublin.

Kohlschütter-Tönz syndrome (KTS), also called Amelo-cerebro-hypohidrotic syndrome is a rare inherited syndrome characterized by epilepsy, psychomotor delay or regression, intellectual disability, and yellow teeth caused by amelogenesis imperfecta. It is a type A ectodermal dysplasia.
A slippery sequence is a small section of codon nucleotide sequences that controls the rate of ribosomal frameshifting. A slippery sequence causes a faster ribosomal transfer which in turn can cause the reading ribosome to "slip." This allows a tRNA to shift by 1 base after it has paired with its anticodon, changing the reading frame.
The bacterial, archaeal and plant plastid code is the DNA code used by bacteria, archaea, prokaryotic viruses and chloroplast proteins. It is essentially the same as the standard code, however there are some variations in alternative start codons.