An inverted repeat is a single stranded sequence of nucleotides followed downstream by its reverse complement. The intervening sequence of nucleotides between the initial sequence and the reverse complement can be any length including zero. For example, 5'---TTACGnnnnnnCGTAA---3' is an inverted repeat sequence. When the intervening length is zero, the composite sequence is a palindromic sequence.

A tumour inducing (Ti) plasmid is a plasmid found in pathogenic species of Agrobacterium, including A. tumefaciens, A. rhizogenes, A. rubi and A. vitis.

Transfer-messenger RNA is a bacterial RNA molecule with dual tRNA-like and messenger RNA-like properties. The tmRNA forms a ribonucleoprotein complex (tmRNP) together with Small Protein B (SmpB), Elongation Factor Tu (EF-Tu), and ribosomal protein S1. In trans-translation, tmRNA and its associated proteins bind to bacterial ribosomes which have stalled in the middle of protein biosynthesis, for example when reaching the end of a messenger RNA which has lost its stop codon. The tmRNA is remarkably versatile: it recycles the stalled ribosome, adds a proteolysis-inducing tag to the unfinished polypeptide, and facilitates the degradation of the aberrant messenger RNA. In the majority of bacteria these functions are carried out by standard one-piece tmRNAs. In other bacterial species, a permuted ssrA gene produces a two-piece tmRNA in which two separate RNA chains are joined by base-pairing.

Stem-loop intramolecular base pairing is a pattern that can occur in single-stranded RNA. The structure is also known as a hairpin or hairpin loop. It occurs when two regions of the same strand, usually complementary in nucleotide sequence when read in opposite directions, base-pair to form a double helix that ends in an unpaired loop. The resulting structure is a key building block of many RNA secondary structures. As an important secondary structure of RNA, it can direct RNA folding, protect structural stability for messenger RNA (mRNA), provide recognition sites for RNA binding proteins, and serve as a substrate for enzymatic reactions.

In molecular biology, the coronavirus frameshifting stimulation element is a conserved stem-loop of RNA found in coronaviruses that can promote ribosomal frameshifting. Such RNA molecules interact with a downstream region to form a pseudoknot structure; the region varies according to the virus but pseudoknot formation is known to stimulate frameshifting. In the classical situation, a sequence 32 nucleotides downstream of the stem is complementary to part of the loop. In other coronaviruses, however, another stem-loop structure around 150 nucleotides downstream can interact with members of this family to form kissing stem-loops and stimulate frameshifting.
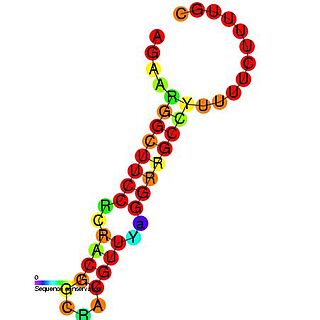
In molecular biology ctRNA is a plasmid encoded noncoding RNA that binds to the mRNA of repB and causes translational inhibition. ctRNA is encoded by plasmids and functions in rolling circle replication to maintain a low copy number. In Corynebacterium glutamicum, it achieves this by antisense pairing with the mRNA of RepB, a replication initiation protein. In Enterococcus faecium the plasmid pJB01 contains three open reading frames, copA, repB, and repC. The pJB01 ctRNA is coded on the opposite strand from the copA/repB intergenic region and partially overlaps an atypical ribosome binding site for repB.
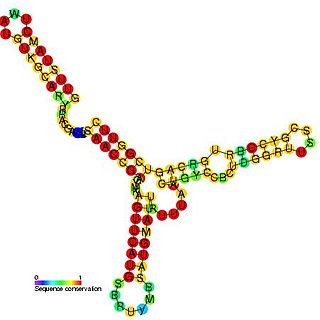
Anti-Q RNA is a small ncRNA from the conjugal plasmid pCF10 of Enterococcus faecalis. It is coded in cis to its regulatory target, prgQ, but can also act in trans. Anti-Q is known to interact with nascent prgQ transcripts to allow formation of an intrinsic terminator, or attenuator, thus preventing transcription of downstream genes. This mode of regulation is essentially the same as that of the countertranscript-driven attenuators that control copy number in pT181, pAMbeta1 and pIP501 and related Staphylococcal plasmids.
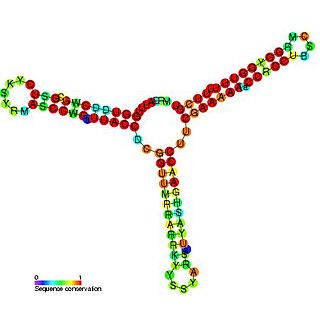
RNAI is a non-coding RNA that is an antisense repressor of the replication of some E. coli plasmids, including ColE1. Plasmid replication is usually initiated by RNAII, which acts as a primer by binding to its template DNA. The complementary RNAI binds RNAII prohibiting it from its initiation role. The rate of degradation of RNAI is therefore a major factor in the control of plasmid replication. This rate of degradation is aided by the pcnB gene product, which polyadenylates the 3' end of RNAI targeting it for degradation by PNPase.

The SAM riboswitch is found upstream of a number of genes which code for proteins involved in methionine or cysteine biosynthesis in Gram-positive bacteria. Two SAM riboswitches in Bacillus subtilis that were experimentally studied act at the level of transcription termination control. The predicted secondary structure consists of a complex stem-loop region followed by a single stem-loop terminator region. An alternative and mutually exclusive form involves bases in the 3' segment of helix 1 with those in the 5' region of helix 5 to form a structure termed the anti-terminator form. When SAM is unbound, the anti-terminator sequence sequesters the terminator sequence so the terminator is unable to form, allowing the polymerase to read-through the downstream gene. When S-Adenosyl methionine (SAM) is bound to the aptamer, the anti-terminator is sequestered by an anti-anti-terminator; the terminator forms and terminates the transcription. However, many SAM riboswitches are likely to regulate gene expression at the level of translation.

The hok/sok system is a postsegregational killing mechanism employed by the R1 plasmid in Escherichia coli. It was the first type I toxin-antitoxin pair to be identified through characterisation of a plasmid-stabilising locus. It is a type I system because the toxin is neutralised by a complementary RNA, rather than a partnered protein.
Ribosomal frameshifting, also known as translational frameshifting or translational recoding, is a biological phenomenon that occurs during translation that results in the production of multiple, unique proteins from a single mRNA. The process can be programmed by the nucleotide sequence of the mRNA and is sometimes affected by the secondary, 3-dimensional mRNA structure. It has been described mainly in viruses, retrotransposons and bacterial insertion elements, and also in some cellular genes.
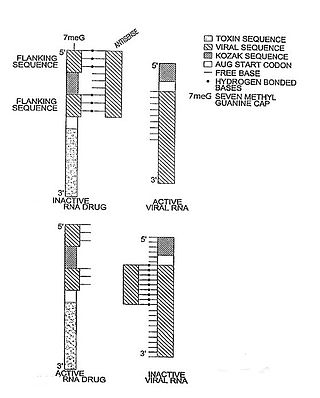
In molecular biology, a riboregulator is a ribonucleic acid (RNA) that responds to a signal nucleic acid molecule by Watson-Crick base pairing. A riboregulator may respond to a signal molecule in any number of manners including, translation of the RNA into a protein, activation of a ribozyme, release of silencing RNA (siRNA), conformational change, and/or binding other nucleic acids. Riboregulators contain two canonical domains, a sensor domain and an effector domain. These domains are also found on riboswitches, but unlike riboswitches, the sensor domain only binds complementary RNA or DNA strands as opposed to small molecules. Because binding is based on base-pairing, a riboregulator can be tailored to differentiate and respond to individual genetic sequences and combinations thereof.

Nucleic acid secondary structure is the basepairing interactions within a single nucleic acid polymer or between two polymers. It can be represented as a list of bases which are paired in a nucleic acid molecule. The secondary structures of biological DNAs and RNAs tend to be different: biological DNA mostly exists as fully base paired double helices, while biological RNA is single stranded and often forms complex and intricate base-pairing interactions due to its increased ability to form hydrogen bonds stemming from the extra hydroxyl group in the ribose sugar.

The regulatory region of the repZ gene, which encodes the replication initiator of plasmid ColIb-P9, contains a pseudoknot. This acts as a molecular switch controlling translation of repZ and repY.
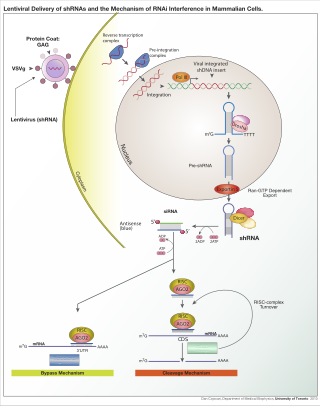
RNA interference (RNAi) is a biological process in which RNA molecules are involved in sequence-specific suppression of gene expression by double-stranded RNA, through translational or transcriptional repression. Historically, RNAi was known by other names, including co-suppression, post-transcriptional gene silencing (PTGS), and quelling. The detailed study of each of these seemingly different processes elucidated that the identity of these phenomena were all actually RNAi. Andrew Fire and Craig C. Mello shared the 2006 Nobel Prize in Physiology or Medicine for their work on RNAi in the nematode worm Caenorhabditis elegans, which they published in 1998. Since the discovery of RNAi and its regulatory potentials, it has become evident that RNAi has immense potential in suppression of desired genes. RNAi is now known as precise, efficient, stable and better than antisense therapy for gene suppression. Antisense RNA produced intracellularly by an expression vector may be developed and find utility as novel therapeutic agents.
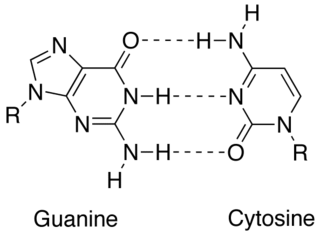
In molecular biology, complementarity describes a relationship between two structures each following the lock-and-key principle. In nature complementarity is the base principle of DNA replication and transcription as it is a property shared between two DNA or RNA sequences, such that when they are aligned antiparallel to each other, the nucleotide bases at each position in the sequences will be complementary, much like looking in the mirror and seeing the reverse of things. This complementary base pairing allows cells to copy information from one generation to another and even find and repair damage to the information stored in the sequences.
Self-complementary adeno-associated virus (scAAV) is a viral vector engineered from the naturally occurring adeno-associated virus (AAV) to be used as a tool for gene therapy. Use of recombinant AAV (rAAV) has been successful in clinical trials addressing a variety of diseases. This lab-made progeny of rAAV is termed "self-complementary" because the coding region has been designed to form an intra-molecular double-stranded DNA template. A rate-limiting step for the standard AAV genome involves the second-strand synthesis since the typical AAV genome is a single-stranded DNA template. However, this is not the case for scAAV genomes. Upon infection, rather than waiting for cell mediated synthesis of the second strand, the two complementary halves of scAAV will associate to form one double stranded DNA (dsDNA) unit that is ready for immediate replication and transcription. The caveat of this construct is that instead of the full coding capacity found in rAAV (4.7–6kb) scAAV can only hold about half of that amount (≈2.4kb).

In genetics, a kissing stem-loop, or kissing stem loop interaction, is formed in ribonucleic acid (RNA) when two bases between two hairpin loops pair. These intra- and intermolecular kissing interactions are important in forming the tertiary or quaternary structure of many RNAs.
In cellular biology, the plasmid copy number is the number of copies of a given plasmid in a cell. To ensure survival and thus the continued propagation of the plasmid, they must regulate their copy number. If a plasmid has too high of a copy number, they may excessively burden their host by occupying too much cellular machinery and using too much energy. On the other hand, too low of a copy number may result in the plasmid not being present in all of their host's progeny. Plasmids may be either low, medium or high copy number plasmids; the regulation mechanisms between low and medium copy number plasmids are different. Low copy plasmids require either a partitioning system or a toxin-antitoxin pair such as CcdA/CcdB to ensure that each daughter receives the plasmid. For example, the F plasmid, which is the origin of BACs is a single copy plasmid with a partitioning system encoded in an operon right next to the plasmid origin. The partitioning system interacts with the septation apparatus to ensure that each daughter receives a copy of the plasmid. Many biotechnology applications utilize mutated plasmids that replicate to high copy number. For example, pBR322 is a medium copy number plasmid from which several high copy number cloning vectors have been derived by mutagenesis, such as the well known pUC series. This delivers the convenience of high plasmid DNA yields but the additional burden of the high copy number restricts the plasmid size. Larger high copy plasmids (>30kb) are disfavoured and also prone to size reduction through deletional mutagenesis.
Coronavirus genomes are positive-sense single-stranded RNA molecules with an untranslated region (UTR) at the 3′ end which is called the 3′ UTR. The 3′ UTR is responsible for important biological functions, such as viral replication. The 3′ UTR has a conserved RNA secondary structure but different Coronavirus genera have different structural features described below.














