
The centromere is the specialized DNA sequence of a chromosome that links a pair of sister chromatids. During mitosis, spindle fibers attach to the centromere via the kinetochore. Centromeres were first thought to be genetic loci that direct the behavior of chromosomes.
Heterochromatin is a tightly packed form of DNA or condensed DNA, which comes in multiple varieties. These varieties lie on a continuum between the two extremes of constitutive heterochromatin and facultative heterochromatin. Both play a role in the expression of genes. Because it is tightly packed, it was thought to be inaccessible to polymerases and therefore not transcribed, however according to Volpe et al. (2002), and many other papers since, much of this DNA is in fact transcribed, but it is continuously turned over via RNA-induced transcriptional silencing (RITS). Recent studies with electron microscopy and OsO4 staining reveal that the dense packing is not due to the chromatin.

Schizosaccharomyces pombe, also called "fission yeast", is a species of yeast used in traditional brewing and as a model organism in molecular and cell biology. It is a unicellular eukaryote, whose cells are rod-shaped. Cells typically measure 3 to 4 micrometres in diameter and 7 to 14 micrometres in length. Its genome, which is approximately 14.1 million base pairs, is estimated to contain 4,970 protein-coding genes and at least 450 non-coding RNAs.

Small interfering RNA (siRNA), sometimes known as short interfering RNA or silencing RNA, is a class of double-stranded RNA non-coding RNA molecules, typically 20-27 base pairs in length, similar to miRNA, and operating within the RNA interference (RNAi) pathway. It interferes with the expression of specific genes with complementary nucleotide sequences by degrading mRNA after transcription, preventing translation.

Dicer, also known as endoribonuclease Dicer or helicase with RNase motif, is an enzyme that in humans is encoded by the DICER1 gene. Being part of the RNase III family, Dicer cleaves double-stranded RNA (dsRNA) and pre-microRNA (pre-miRNA) into short double-stranded RNA fragments called small interfering RNA and microRNA, respectively. These fragments are approximately 20-25 base pairs long with a two-base overhang on the 3' ends. Dicer facilitates the activation of the RNA-induced silencing complex (RISC), which is essential for RNA interference. RISC has a catalytic component Argonaute, which is an endonuclease capable of degrading messenger RNA (mRNA).
The RNA-induced silencing complex, or RISC, is a multiprotein complex, specifically a ribonucleoprotein, which functions in gene silencing via a variety of pathways at the transcriptional and translational levels. Using single-stranded RNA (ssRNA) fragments, such as microRNA (miRNA), or double-stranded small interfering RNA (siRNA), the complex functions as a key tool in gene regulation. The single strand of RNA acts as a template for RISC to recognize complementary messenger RNA (mRNA) transcript. Once found, one of the proteins in RISC, Argonaute, activates and cleaves the mRNA. This process is called RNA interference (RNAi) and it is found in many eukaryotes; it is a key process in defense against viral infections, as it is triggered by the presence of double-stranded RNA (dsRNA).
Subtelomeres are segments of DNA between telomeric caps and chromatin.
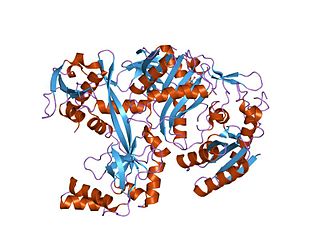
Ribonuclease III (BRENDA 3.1.26.3) is a type of ribonuclease that recognizes dsRNA and cleaves it at specific targeted locations to transform them into mature RNAs. These enzymes are a group of endoribonucleases that are characterized by their ribonuclease domain, which is labelled the RNase III domain. They are ubiquitous compounds in the cell and play a major role in pathways such as RNA precursor synthesis, RNA Silencing, and the pnp autoregulatory mechanism.

The Argonaute protein family plays a central role in RNA silencing processes, as essential components of the RNA-induced silencing complex (RISC). RISC is responsible for the gene silencing phenomenon known as RNA interference (RNAi). Argonaute proteins bind different classes of small non-coding RNAs, including microRNAs (miRNAs), small interfering RNAs (siRNAs) and Piwi-interacting RNAs (piRNAs). Small RNAs guide Argonaute proteins to their specific targets through sequence complementarity, which then leads to mRNA cleavage or translation inhibition.

A chromodomain is a protein structural domain of about 40–50 amino acid residues commonly found in proteins associated with the remodeling and manipulation of chromatin. The domain is highly conserved among both plants and animals, and is represented in a large number of different proteins in many genomes, such as that of the mouse. Some chromodomain-containing genes have multiple alternative splicing isoforms that omit the chromodomain entirely. In mammals, chromodomain-containing proteins are responsible for aspects of gene regulation related to chromatin remodeling and formation of heterochromatin regions. Chromodomain-containing proteins also bind methylated histones and appear in the RNA-induced transcriptional silencing complex. In histone modifications, chromodomains are very conserved. They function by identifying and binding to methylated lysine residues that exist on the surface of chromatin proteins and thereby regulate gene transcription.
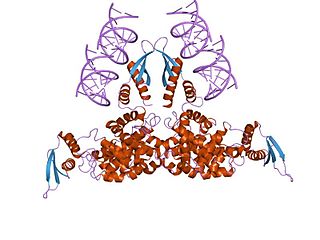
Piwi genes were identified as regulatory proteins responsible for stem cell and germ cell differentiation. Piwi is an abbreviation of P-elementInduced WImpy testis in Drosophila. Piwi proteins are highly conserved RNA-binding proteins and are present in both plants and animals. Piwi proteins belong to the Argonaute/Piwi family and have been classified as nuclear proteins. Studies on Drosophila have also indicated that Piwi proteins have slicer activity conferred by the presence of the Piwi domain. In addition, Piwi associates with heterochromatin protein 1, an epigenetic modifier, and piRNA-complementary sequences. These are indications of the role Piwi plays in epigenetic regulation. Piwi proteins are also thought to control the biogenesis of piRNA as many Piwi-like proteins contain slicer activity which would allow Piwi proteins to process precursor piRNA into mature piRNA.
RNA-induced transcriptional silencing (RITS) is a form of RNA interference by which short RNA molecules – such as small interfering RNA (siRNA) – trigger the downregulation of transcription of a particular gene or genomic region. This is usually accomplished by posttranslational modification of histone tails which target the genomic region for heterochromatin formation. The protein complex that binds to siRNAs and interacts with the methylated lysine 9 residue of histones H3 is the RITS complex.
The mating-type region is a specialized region in the genomes of some yeast and other fungi, usually organized into heterochromatin and possessing unique histone methylation patterns. The genes in this region regulate the mating type of the organism and therefore determine key events in its life cycle, such as whether it will reproduce sexually or asexually. In fission yeast such as S. pombe, the formation and maintenance of the heterochromatin organization is regulated by RNA-induced transcriptional silencing, a form of RNA interference responsible for genomic maintenance in many organisms. Mating type regions have also been well studied in budding yeast S. cerevisiae and in the fungus Neurospora crassa.
RNA silencing or RNA interference refers to a family of gene silencing effects by which gene expression is negatively regulated by non-coding RNAs such as microRNAs. RNA silencing may also be defined as sequence-specific regulation of gene expression triggered by double-stranded RNA (dsRNA). RNA silencing mechanisms are highly conserved in most eukaryotes. The most common and well-studied example is RNA interference (RNAi), in which endogenously expressed microRNA (miRNA) or exogenously derived small interfering RNA (siRNA) induces the degradation of complementary messenger RNA. Other classes of small RNA have been identified, including piwi-interacting RNA (piRNA) and its subspecies repeat associated small interfering RNA (rasiRNA).

Centromere protein H is a protein that in humans is encoded by the CENPH gene.
RNA polymerase IV is an enzyme that synthesizes small interfering RNA (siRNA) in plants, which silence gene expression. RNAP IV belongs to a family of enzymes that catalyze the process of transcription known as RNA Polymerases, which synthesize RNA from DNA templates. Discovered via phylogenetic studies of land plants, genes of RNAP IV are thought to have resulted from multistep evolution processes that occurred in RNA Polymerase II phylogenies. Such an evolutionary pathway is supported by the fact that RNAP IV is composed of 12 protein subunits that are either similar or identical to RNA polymerase II, and is specific to plant genomes. Via its synthesis of siRNA, RNAP IV is involved in regulation of heterochromatin formation in a process known as RNA directed DNA Methylation (RdDM).

RNA interference (RNAi) is a biological process in which RNA molecules are involved in sequence-specific suppression of gene expression by double-stranded RNA, through translational or transcriptional repression. Historically, RNAi was known by other names, including co-suppression, post-transcriptional gene silencing (PTGS), and quelling. The detailed study of each of these seemingly different processes elucidated that the identity of these phenomena were all actually RNAi. Andrew Fire and Craig C. Mello shared the 2006 Nobel Prize in Physiology or Medicine for their work on RNA interference in the nematode worm Caenorhabditis elegans, which they published in 1998. Since the discovery of RNAi and its regulatory potentials, it has become evident that RNAi has immense potential in suppression of desired genes. RNAi is now known as precise, efficient, stable and better than antisense therapy for gene suppression. Antisense RNA produced intracellularly by an expression vector may be developed and find utility as novel therapeutic agents.

Telomeric repeat-containing RNA (TERRA) is a long non-coding RNA transcribed from telomeres - repetitive nucleotide regions found on the ends of chromosomes that function to protect DNA from deterioration or fusion with neighboring chromosomes. TERRA has been shown to be ubiquitously expressed in almost all cell types containing linear chromosomes - including humans, mice, and yeasts. While the exact function of TERRA is still an active area of research, it is generally believed to play a role in regulating telomerase activity as well as maintaining the heterochromatic state at the ends of chromosomes. TERRA interaction with other associated telomeric proteins has also been shown to help regulate telomere integrity in a length-dependent manner.
Robin Campbell Allshire is Professor of Chromosome Biology at University of Edinburgh and a Wellcome Trust Principal Research Fellow. His research group at the Wellcome Trust Centre for Cell Biology focuses on the epigenetic mechanisms governing the assembly of specialised domains of chromatin and their transmission through cell division.

RNA-directed DNA methylation (RdDM) is a biological process in which non-coding RNA molecules direct the addition of DNA methylation to specific DNA sequences. The RdDM pathway is unique to plants, although other mechanisms of RNA-directed chromatin modification have also been described in fungi and animals. To date, the RdDM pathway is best characterized within angiosperms, and particularly within the model plant Arabidopsis thaliana. However, conserved RdDM pathway components and associated small RNAs (sRNAs) have also been found in other groups of plants, such as gymnosperms and ferns. The RdDM pathway closely resembles other sRNA pathways, particularly the highly conserved RNAi pathway found in fungi, plants, and animals. Both the RdDM and RNAi pathways produce sRNAs and involve conserved Argonaute, Dicer and RNA-dependent RNA polymerase proteins.












