
Ribosomes are macromolecular machines, found within all living cells, that perform biological protein synthesis. Ribosomes link amino acids together in the order specified by the codons of messenger RNA (mRNA) molecules to form polypeptide chains. Ribosomes consist of two major components: the small and large ribosomal subunits. Each subunit consists of one or more ribosomal RNA (rRNA) molecules and many ribosomal proteins. The ribosomes and associated molecules are also known as the translational apparatus.

In molecular biology, a riboswitch is a regulatory segment of a messenger RNA molecule that binds a small molecule, resulting in a change in production of the proteins encoded by the mRNA. Thus, an mRNA that contains a riboswitch is directly involved in regulating its own activity, in response to the concentrations of its effector molecule. The discovery that modern organisms use RNA to bind small molecules, and discriminate against closely related analogs, expanded the known natural capabilities of RNA beyond its ability to code for proteins, catalyze reactions, or to bind other RNA or protein macromolecules.

A ribosomal protein is any of the proteins that, in conjunction with rRNA, make up the ribosomal subunits involved in the cellular process of translation. E. coli, other bacteria and Archaea have a 30S small subunit and a 50S large subunit, whereas humans and yeasts have a 40S small subunit and a 60S large subunit. Equivalent subunits are frequently numbered differently between bacteria, Archaea, yeasts and humans.

This family is a putative ribosomal protein leader autoregulatory structure found in B. subtilis and other low-GC Gram-positive bacteria. It is located in the 5′ untranslated regions of mRNAs encoding ribosomal proteins L10 and L12 (rplJ-rplL). A Rho-independent transcription terminator structure that is probably involved in regulation is included at the 3′ end.
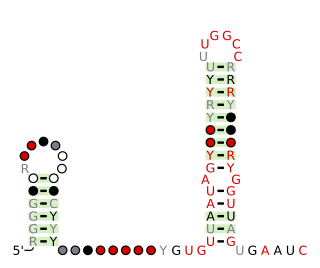
L13 ribosomal protein leaders are part of the ribosome biogenesis. They are used as an autoregulatory mechanism to control the concentration of ribosomal proteins L13. An experimentally confirmed ribosomal protein leader autoregulatory structure was found in B. subtilis and other low-GC Gram-positive bacteria. It is located in the 5′ untranslated regions of mRNAs encoding the L13-S9 (rplM-rpsI) ribosomal protein operon. A second example were predicted in Bacteroidia with bioinformatic approaches, but the structure of the putative Bacteroidia example is apparently unrelated to the previously established L13 ribosomal protein leader.

L20 ribosomal protein leader is a ribosomal protein leader involved in the ribosome biogenesis. It is used as an autoregulatory mechanism to control the concentration of ribosomal proteins L20. The structure is located in the 5′ untranslated regions of mRNAs encoding initiation factor 3 followed by ribosomal proteins L35 and L20 (infC-rpmI-rplT). A Rho-independent transcription terminator structure that is probably involved in regulation is included at the 3′ end. This family is a putative ribosomal protein leader autoregulatory structure found in B. subtilis and other low-GC Gram-positive bacteria. A second example were predicted in Deltaproteobacteria with bioinformatic approaches. In this structure were similarities between the rRNA binding site of the L20 ribosomal protein and the L20 ribosomal protein leader binding site detected.

This family is a putative ribosomal protein leader autoregulatory structure found in B. subtilis and other low-GC Gram-positive bacteria. It is located in the 5′ untranslated regions of mRNAs encoding ribosomal protein L21, a protein of unknown function, and ribosomal protein L27 (rplU-ysxB-rpmA).

S15 Ribosomal protein leaders are part of the ribosome biogenesis. They were used as an autoregulatory mechanism to control the concentration of ribosomal proteins S15. The structure is located in the 5′ untranslated regions of mRNAs encoding ribosomal proteins S15 (rpsO). Multiple distinct structural types of S15 ribosomal protein leaders are known in different organisms.

The 23S methyl RNA motif is a conserved RNA structure found upstream of genes predicted to encode rRNA methyltransferases, possibly for 23S rRNA. However, in one case it is far from the rRNA methyltransferase gene. Nonetheless, it was proposed that this RNA could be a cis-regulatory element, an attractive hypothesis in view of the fact that rRNA methyltransferases can bind RNA, and therefore presumably the 23S methyl RNA. This regulatory strategy is common in bacteria to control levels of ribosomal subunits, using RNA motifs called ribosomal leaders. Additionally, 23S methyl RNAs have rho-independent transcription terminators downstream, which could be a mechanism of regulation.

The L17 downstream element RNA motif is a conserved RNA structure identified in bacteria by bioinformatics. All known L17 downstream elements were detected immediately downstream of genes encoding the L17 subunit of the ribosome, and therefore might be in the 3' untranslated regions of these genes. The element is found in a variety of lactic acid bacteria and in the genus Listeria.

The L2 ribosomal protein leader is a ribosomal protein leader involved in ribosome biogenesis. It is used as an autoregulatory mechanism to control the concentration of the ribosomal protein L2. Known Examples were predicted in Alphaproteobacteria with bioinformatic approaches. The structure is located in the 5′ untranslated regions of mRNAs encoding ribosomal proteins L2 (rplB), S30 (rpsS), L22 (rplV) and S3 (rpsC). It was proposed that the ligand is uncertain, because none of the downstream (regulated) genes are known as a previously established ribosomal protein leader ligand.

L25 ribosomal protein leader is a ribosomal protein leader involved in the ribosome biogenesis. It is used as an autoregulatory mechanism to control the concentration of the ribosomal protein L25. Known Examples were predicted in Gammaproteobacteria with bioinformatic approaches. or in Enterobacteria. The structure is located in the 5′ untranslated regions of mRNAs encoding ribosomal protein L25 (rplY).

S10 ribosomal protein leader is a ribosomal protein leader involved in the ribosome biogenesis. It is used as an autoregulatory mechanism to control the concentration of the ribosomal protein S10. Known Examples were predicted in Clostridia or other lineages of Firmicutes with bioinformatic approaches. The structure is located in the 5′ untranslated regions of mRNAs encoding ribosomal proteins S10 (rpsJ), L3 (rplc) and L4 (rplD). There is an uncertainty about the ligand, because of a lack of experimental investigation.

The S4 ribosomal protein leader is a ribosomal protein leader involved in ribosome biogenesis. It is used as an autoregulatory mechanism to control the concentration of the ribosomal protein S4. Two examples of such leaders that use different conserved structures, in Firmicutes and Gammaproteobacteria, have been experimentally confirmed. Four additional S4 ribosomal protein leaders, each with distinct structures, were predicted in various bacteria phyla. In Bacteroidia or Firmicutes, the structure is located in the 5′ untranslated regions of mRNAs encoding ribosomal proteins S4 (rpsD), RNA polymerase alpha subunit (rpoA) and L17 (rplQ). In Clostridia and Gammaproteobacteria, the ribosomal proteins S13 (rpsM) and S11 (rpsK) were also part of the mRNA encoding region.

An eL15 ribosomal protein leader is a ribosomal protein leader involved in ribosome biogenesis. It is used as an autoregulatory mechanism to control the concentration of the ribosomal protein eL15, which is used in archaea and eukaryotes. Known Examples were predicted in Euryarchaeota with bioinformatic approaches. The structure is located in the 5′ untranslated regions of mRNAs encoding ribosomal proteins eL15 (rpl15e). Similarities between the eL15 ribosomal protein and the rRNA site to which this protein binds were detected.

An L17 ribosomal protein leader is a ribosomal protein leader involved in ribosome biogenesis. It is used as an autoregulatory mechanism to control the concentration of the ribosomal protein L17. Known Examples were predicted in Actinobacteria and Proteobacteria with bioinformatic approaches. The structure is located in the 5′ untranslated regions of mRNAs encoding ribosomal protein L17 (rplQ).

An L31 ribosomal protein leader is a ribosomal protein leader involved in ribosome biogenesis. It is used as an autoregulatory mechanism to control the concentration of the ribosomal protein L31. Five structurally distinct types of L31 ribosomal protein leader were predicted with a bioinformatic approach. The structure is located in the 5′ untranslated regions of mRNAs encoding ribosomal protein L31 (rpmE), and in one case L32 (rpmF). These are found in different species of Actinobacteria, Firmicutes or Gammaproteobacteria. The gammaproteobacterial type was also detected and validated in an independent experimental study using the organism Escherichia coli.

A S16 ribosomal protein leader is a ribosomal protein leader involved in ribosome biogenesis. It is used as an autoregulatory mechanism to control the concentration of the ribosomal protein S16. Known Examples were predicted in Flavobacteria with bioinformatic approaches. The structure is located in the 5′ untranslated regions of mRNAs encoding ribosomal proteins L16 (rpsP) and the ribosome maturation factor protein (rimM).

S6:S18 ribosomal protein leader is a ribosomal protein leader involved in the ribosome biogenesis. It is used as an autoregulatory mechanism to control the concentration of the ribosomal proteins S6:S18 complex. An experimentally confirmed example of such a leader occurs in a wide variety of bacteria, though not all phyla. A S6:S18 ribosomal leader was predicted in Chlorobia, and its predicted structure differs from that of the validated S6:S18 ribosomal leader. This structure is located in the 5′ untranslated regions of mRNAs encoding ribosomal proteins rpsF (S6), the Single-strand DNA-binding protein A (ssbA), S18 (rpsR) and L7/L12 (rpll).
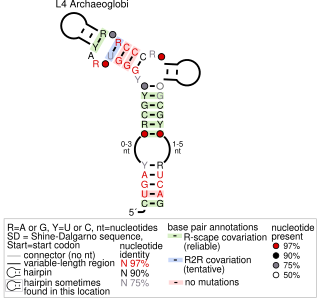
An L4 ribosomal protein leader is a ribosomal protein leader involved in ribosome biogenesis. It is used as an autoregulatory mechanism to control the concentration of the ribosomal protein L4. Known Examples were predicted in Archaeoglobi with bioinformatic approaches. The structure is located in the 5′ untranslated regions of mRNAs encoding ribosomal proteins L3 (rplC), L4 (rplD), L23 (rplW) and L2 (rplB).





















