Related Research Articles
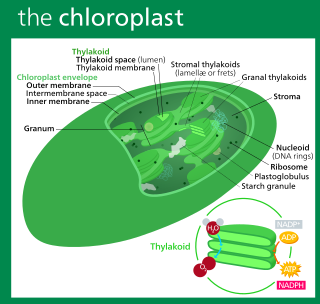
A chloroplast is a type of membrane-bound organelle known as a plastid that conducts photosynthesis mostly in plant and algal cells. The photosynthetic pigment chlorophyll captures the energy from sunlight, converts it, and stores it in the energy-storage molecules ATP and NADPH while freeing oxygen from water in the cells. The ATP and NADPH is then used to make organic molecules from carbon dioxide in a process known as the Calvin cycle. Chloroplasts carry out a number of other functions, including fatty acid synthesis, amino acid synthesis, and the immune response in plants. The number of chloroplasts per cell varies from one, in unicellular algae, up to 100 in plants like Arabidopsis and wheat.
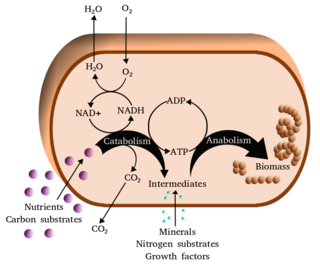
Metabolism is the set of life-sustaining chemical reactions in organisms. The three main functions of metabolism are: the conversion of the energy in food to energy available to run cellular processes; the conversion of food to building blocks of proteins, lipids, nucleic acids, and some carbohydrates; and the elimination of metabolic wastes. These enzyme-catalyzed reactions allow organisms to grow and reproduce, maintain their structures, and respond to their environments. The word metabolism can also refer to the sum of all chemical reactions that occur in living organisms, including digestion and the transportation of substances into and between different cells, in which case the above described set of reactions within the cells is called intermediary metabolism.

Photosynthesis is a system of biological processes by which photosynthetic organisms, such as most plants, algae, and cyanobacteria, convert light energy, typically from sunlight, into the chemical energy necessary to fuel their activities. Photosynthetic organisms use intracellular organic compounds to store the chemical energy they produce in photosynthesis within organic compounds like sugars, glycogen, cellulose and starches. Photosynthesis is usually used to refer to oxygenic photosynthesis, a process that produces oxygen. To use this stored chemical energy, the organisms' cells metabolize the organic compounds through another process called cellular respiration. Photosynthesis plays a critical role in producing and maintaining the oxygen content of the Earth's atmosphere, and it supplies most of the biological energy necessary for complex life on Earth.
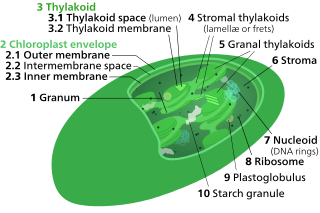
Thylakoids are membrane-bound compartments inside chloroplasts and cyanobacteria. They are the site of the light-dependent reactions of photosynthesis. Thylakoids consist of a thylakoid membrane surrounding a thylakoid lumen. Chloroplast thylakoids frequently form stacks of disks referred to as grana. Grana are connected by intergranal or stromal thylakoids, which join granum stacks together as a single functional compartment.

Chloroplasts contain several important membranes, vital for their function. Like mitochondria, chloroplasts have a double-membrane envelope, called the chloroplast envelope, but unlike mitochondria, chloroplasts also have internal membrane structures called thylakoids. Furthermore, one or two additional membranes may enclose chloroplasts in organisms that underwent secondary endosymbiosis, such as the euglenids and chlorarachniophytes.

Photorespiration (also known as the oxidative photosynthetic carbon cycle or C2 cycle) refers to a process in plant metabolism where the enzyme RuBisCO oxygenates RuBP, wasting some of the energy produced by photosynthesis. The desired reaction is the addition of carbon dioxide to RuBP (carboxylation), a key step in the Calvin–Benson cycle, but approximately 25% of reactions by RuBisCO instead add oxygen to RuBP (oxygenation), creating a product that cannot be used within the Calvin–Benson cycle. This process lowers the efficiency of photosynthesis, potentially lowering photosynthetic output by 25% in C3 plants. Photorespiration involves a complex network of enzyme reactions that exchange metabolites between chloroplasts, leaf peroxisomes and mitochondria.
Gibberellins (GAs) are plant hormones that regulate various developmental processes, including stem elongation, germination, dormancy, flowering, flower development, and leaf and fruit senescence. GAs are one of the longest-known classes of plant hormone. It is thought that the selective breeding of crop strains that were deficient in GA synthesis was one of the key drivers of the "green revolution" in the 1960s, a revolution that is credited to have saved over a billion lives worldwide.

Jasmonate (JA) and its derivatives are lipid-based plant hormones that regulate a wide range of processes in plants, ranging from growth and photosynthesis to reproductive development. In particular, JAs are critical for plant defense against herbivory and plant responses to poor environmental conditions and other kinds of abiotic and biotic challenges. Some JAs can also be released as volatile organic compounds (VOCs) to permit communication between plants in anticipation of mutual dangers.

Indole-3-acetic acid is the most common naturally occurring plant hormone of the auxin class. It is the best known of the auxins, and has been the subject of extensive studies by plant physiologists. IAA is a derivative of indole, containing a carboxymethyl substituent. It is a colorless solid that is soluble in polar organic solvents.
Metabolic network modelling, also known as metabolic network reconstruction or metabolic pathway analysis, allows for an in-depth insight into the molecular mechanisms of a particular organism. In particular, these models correlate the genome with molecular physiology. A reconstruction breaks down metabolic pathways into their respective reactions and enzymes, and analyzes them within the perspective of the entire network. In simplified terms, a reconstruction collects all of the relevant metabolic information of an organism and compiles it in a mathematical model. Validation and analysis of reconstructions can allow identification of key features of metabolism such as growth yield, resource distribution, network robustness, and gene essentiality. This knowledge can then be applied to create novel biotechnology.
Systemin is a plant peptide hormone involved in the wound response in the family Solanaceae. It was the first plant hormone that was proven to be a peptide having been isolated from tomato leaves in 1991 by a group led by Clarence A. Ryan. Since then, other peptides with similar functions have been identified in tomato and outside of the Solanaceae. Hydroxyproline-rich glycopeptides were found in tobacco in 2001 and AtPeps were found in Arabidopsis thaliana in 2006. Their precursors are found both in the cytoplasm and cell walls of plant cells, upon insect damage, the precursors are processed to produce one or more mature peptides. The receptor for systemin was first thought to be the same as the brassinolide receptor but this is now uncertain. The signal transduction processes that occur after the peptides bind are similar to the cytokine-mediated inflammatory immune response in animals. Early experiments showed that systemin travelled around the plant after insects had damaged the plant, activating systemic acquired resistance, now it is thought that it increases the production of jasmonic acid causing the same result. The main function of systemins is to coordinate defensive responses against insect herbivores but they also affect plant development. Systemin induces the production of protease inhibitors which protect against insect herbivores, other peptides activate defensins and modify root growth. They have also been shown to affect plants' responses to salt stress and UV radiation. AtPEPs have been shown to affect resistance against oomycetes and may allow A. thaliana to distinguish between different pathogens. In Nicotiana attenuata, some of the peptides have stopped being involved in defensive roles and instead affect flower morphology.

An aromatic amino acid is an amino acid that includes an aromatic ring.

12-oxophytodienoate reductase (OPRs) is an enzyme of the family of Old Yellow Enzymes (OYE). OPRs are grouped into two groups: OPRI and OPRII – the second group is the focus of this article, as the function of the first group is unknown, but is the subject of current research. The OPR enzyme utilizes the cofactor flavin mononucleotide (FMN) and catalyzes the following reaction in the jasmonic acid synthesis pathway:
In enzymology, an L-2-hydroxyglutarate dehydrogenase is an enzyme that catalyzes the chemical reaction
In enzymology, a ferredoxin-NADP+ reductase (EC 1.18.1.2) abbreviated FNR, is an enzyme that catalyzes the chemical reaction
Peptide signaling plays a significant role in various aspects of plant growth and development and specific receptors for various peptides have been identified as being membrane-localized receptor kinases, the largest family of receptor-like molecules in plants. Signaling peptides include members of the following protein families.
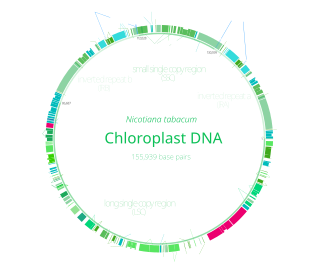
Chloroplast DNA (cpDNA) is the DNA located in chloroplasts, which are photosynthetic organelles located within the cells of some eukaryotic organisms. Chloroplasts, like other types of plastid, contain a genome separate from that in the cell nucleus. The existence of chloroplast DNA was identified biochemically in 1959, and confirmed by electron microscopy in 1962. The discoveries that the chloroplast contains ribosomes and performs protein synthesis revealed that the chloroplast is genetically semi-autonomous. The first complete chloroplast genome sequences were published in 1986, Nicotiana tabacum (tobacco) by Sugiura and colleagues and Marchantia polymorpha (liverwort) by Ozeki et al. Since then, a great number of chloroplast DNAs from various species have been sequenced.
In molecular biology, the PsbZ (Ycf9) is a protein domain, which is low in molecular weight. It is a transmembrane protein and therefore is located in the thylakoid membrane of chloroplasts in cyanobacteria and plants. More specifically, it is located in Photosystem II (PSII) and in the light-harvesting complex II (LHCII). Ycf9 acts as a structural linker, that stabilises the PSII-LHCII supercomplexes. Moreover, the supercomplex fails to form in PsbZ-deficient mutants, providing further evidence to suggest Ycf9's role as a structural linker. This may be caused by a marked decrease in two LHCII antenna proteins, CP26 and CP29, found in PsbZ-deficient mutants, which result in structural changes, as well as functional modifications in PSII.

Chlororespiration is a respiratory process that takes place within plants. Inside plant cells there is an organelle called the chloroplast which is surrounded by the thylakoid membrane. This membrane contains an enzyme called NAD(P)H dehydrogenase which transfers electrons in a linear chain to oxygen molecules. This electron transport chain (ETC) within the chloroplast also interacts with those in the mitochondria where respiration takes place. Photosynthesis is also a process that Chlororespiration interacts with. If photosynthesis is inhibited by environmental stressors like water deficit, increased heat, and/or increased/decreased light exposure, or even chilling stress then chlororespiration is one of the crucial ways that plants use to compensate for chemical energy synthesis.
Plastid terminal oxidase or plastoquinol terminal oxidase (PTOX) is an enzyme that resides on the thylakoid membranes of plant and algae chloroplasts and on the membranes of cyanobacteria. The enzyme was hypothesized to exist as a photosynthetic oxidase in 1982 and was verified by sequence similarity to the mitochondrial alternative oxidase (AOX). The two oxidases evolved from a common ancestral protein in prokaryotes, and they are so functionally and structurally similar that a thylakoid-localized AOX can restore the function of a PTOX knockout.
References
- 1 2 3 4 "Robert L. Last". msu.edu. Retrieved August 26, 2017.
- ↑ "Robert L. Last" . Retrieved August 26, 2017.
- ↑ "CV" (PDF). cornell.edu. Retrieved August 26, 2017.
- 1 2 Jander, G; et al. (2002). "Arabidopsis map-based cloning in the post-genome era". Plant Physiology. 129 (2): 440–450. doi:10.1104/pp.003533. PMC 1540230 . PMID 12068090.
- ↑ Schiavo, Fiorella Lo; Last, Robert L.; Morelli, Giorgio; Raikhel, Natasha V. (29 June 2013). Cellular Integration of Signalling Pathways in Plant Development. ISBN 9783642721175 . Retrieved August 26, 2017.
- ↑ Last, RL; Fink, GR (1988). "Tryptophan-requiring mutants of the plant Arabidopsis thaliana". Science. 240 (4850): 305–310. Bibcode:1988Sci...240..305L. doi:10.1126/science.240.4850.305. PMID 17796738. S2CID 39917514.
- ↑ Radwanski, ER; Last, RL (1995). "Tryptophan biosynthesis and metabolism: Biochemical and molecular genetics". Plant Cell. 7 (7): 921–934. doi:10.2307/3870047. JSTOR 3870047. PMC 160888 . PMID 7640526.
- ↑ Gu, L (2010). "Metabolite profiling reveals broad metabolic phenotypes associated with a plant amino acid catabolism mutant". Plant Journal. 61 (4): 579–590. doi: 10.1111/j.1365-313x.2009.04083.x . PMID 19929878.
- ↑ Van Eenennaam, AL (2003). "Engineering improved vitamin E quality: from Arabidopsis mutant to soy oil". Plant Cell. 15 (12): 3007–3019. doi: 10.1105/tpc.015875 . PMC 282849 . PMID 14630966.
- ↑ Li, J; et al. (1993). "Arabidopsis flavonoid mutants are hypersensitive to UV-B irradiation". Plant Cell. 5 (2): 171–179. doi:10.2307/3869583. JSTOR 3869583. PMC 160260 . PMID 12271060.
- ↑ Landry, LG; et al. (1997). "An Arabidopsis photolyase mutant is hypersensitive to ultraviolet-B radiation". Proc. Natl. Acad. Sci. USA. 94 (1): 328–332. Bibcode:1997PNAS...94..328L. doi: 10.1073/pnas.94.1.328 . PMC 19334 . PMID 8990208.
- ↑ Kliebenstein, DJ; et al. (2002). "The Arabidopsis RCC1 homologue UVR8 mediates UV-B signal transduction and tolerance". Plant Physiology. 130 (1): 234–243. doi:10.1104/pp.005041. PMC 166556 . PMID 12226503.
- ↑ Lu, Y; et al. (2011). "A small zinc finger thylakoid protein plays a role in maintenance of photosystem II". Plant Cell. 23 (5): 1861–1875. doi: 10.1105/tpc.111.085456 . PMC 3123961 . PMID 21586683.
- ↑ Liu, Jun; Last, Robert L. (2017-09-19). "A chloroplast thylakoid lumen protein is required for proper photosynthetic acclimation of plants under fluctuating light environments". Proceedings of the National Academy of Sciences. 114 (38): E8110–E8117. Bibcode:2017PNAS..114E8110L. doi: 10.1073/pnas.1712206114 . ISSN 0027-8424. PMC 5617312 . PMID 28874535.
- ↑ Schilmiller, AL; Schauvinhold, I; et al. (2009). "Monoterpenes in the glandular trichomes of tomato are synthesized via a neryl diphosphate intermediate rather than geranyl diphosphate". Proc. Natl. Acad. Sci. USA. 106 (26): 10865–70. doi: 10.1073/pnas.0904113106 . PMC 2705607 . PMID 19487664.
- ↑ Milo, R; Last, RL (2012). "Achieving diversity in the face of constraints - lessons from metabolism". Science. 336 (6089): 1663–1667. Bibcode:2012Sci...336.1663M. doi:10.1126/science.1217665. PMID 22745419. S2CID 206539296.
- ↑ Schilmiller, AL; et al. (2012). "Identification of a BAHD acetyltransferase that produces protective acyl sugars in tomato trichomes". Proc. Natl. Acad. Sci. USA. 109 (40): 16377–16382. doi: 10.1073/pnas.1207906109 . PMC 3479610 . PMID 22988115.
- ↑ Liu, J; Last, RL (2015). ". A land plant-specific thylakoid membrane protein contributes to photosystem II maintenance in Arabidopsis thaliana". Plant Journal. 82 (5): 731–743. doi: 10.1111/tpj.12845 . PMID 25846821.
- ↑ Fan, P; et al. (2016). "In vitro reconstruction and analysis of evolutionary variation of the tomato acylsucrose metabolic network". Proc. Natl. Acad. Sci. USA. 113 (2): E239-48. Bibcode:2016PNAS..113E.239F. doi: 10.1073/pnas.1517930113 . PMC 4720351 . PMID 26715757.
- ↑ Fan, P; Miller, AM; Liu, X; Jones, AD; Last, RL (2017). "Evolution of a flipped pathway creates metabolic innovation in tomato trichomes through BAHD enzyme promiscuity". Nature Communications. 8 (1): 2080. Bibcode:2017NatCo...8.2080F. doi:10.1038/s41467-017-02045-7. PMC 5727100 . PMID 29234041.
- ↑ Moghe, GD; Leong, BJ; Hurney, SM; Jones, AD; Last, RL (2017). "Evolutionary routes to biochemical innovation revealed by integrative analysis of a plant-defense related specialized metabolic pathway". eLife. 6: e38468. doi: 10.7554/eLife.28468 . PMC 5595436 . PMID 28853706.
- ↑ Leong, Bryan J.; Lybrand, Daniel B.; Lou, Yann-Ru; Fan, Pengxiang; Schilmiller, Anthony L.; Last, Robert L. (2019-04-01). "Evolution of metabolic novelty: A trichome-expressed invertase creates specialized metabolic diversity in wild tomato". Science Advances. 5 (4): eaaw3754. Bibcode:2019SciA....5.3754L. doi: 10.1126/sciadv.aaw3754 . ISSN 2375-2548. PMC 6482016 . PMID 31032420.