
Human metapneumovirus is a negative-sense single-stranded RNA virus of the family Pneumoviridae and is closely related to the Avian metapneumovirus (AMPV) subgroup C. It was isolated for the first time in 2001 in the Netherlands by using the RAP-PCR technique for identification of unknown viruses growing in cultured cells. As of 2016, it was the second most common cause of acute respiratory tract illness in otherwise-healthy children under the age of 5 in a large US outpatient clinic.

Influenza hemagglutinin (HA) or haemagglutinin[p] is a homotrimeric glycoprotein found on the surface of influenza viruses and is integral to its infectivity.

Orthomyxoviridae is a family of negative-sense RNA viruses. It includes seven genera: Alphainfluenzavirus, Betainfluenzavirus, Gammainfluenzavirus, Deltainfluenzavirus, Isavirus, Thogotovirus, and Quaranjavirus. The first four genera contain viruses that cause influenza in birds and mammals, including humans. Isaviruses infect salmon; the thogotoviruses are arboviruses, infecting vertebrates and invertebrates. The Quaranjaviruses are also arboviruses, infecting vertebrates (birds) and invertebrates (arthropods).

Respiratory tract infections (RTIs) are infectious diseases involving the lower or upper respiratory tract. An infection of this type usually is further classified as an upper respiratory tract infection or a lower respiratory tract infection. Lower respiratory infections, such as pneumonia, tend to be far more severe than upper respiratory infections, such as the common cold.

Alphacoronavirus amsterdamense is a species of coronavirus, specifically a Setracovirus from among the Alphacoronavirus genus. It was identified in late 2004 in patients in the Netherlands by Lia van der Hoek and Krzysztof Pyrc using a novel virus discovery method VIDISCA. Later on the discovery was confirmed by the researchers from Rotterdam. The virus is an enveloped, positive-sense, single-stranded RNA virus which enters its host cell by binding to ACE2. Infection with the virus has been confirmed worldwide, and has an association with many common symptoms and diseases. Associated diseases include mild to moderate upper respiratory tract infections, severe lower respiratory tract infection, croup and bronchiolitis.

Influenza B virus is the only species in the genus Betainfluenzavirus in the virus family Orthomyxoviridae.

Influenza A virus subtype H10N7 (A/H10N7) is a subtype of the species Influenza A virus. H10N7 was first reported in humans in Egypt in 2004. It caused illness in two one-year-old infants, and residents of Ismailia, Egypt; one child's father, and a poultry merchant.
Immune stimulating complexes (ISCOMs) are spherical open cage-like structures (typically 40 nm in diameter) that are spontaneously formed when mixing together cholesterol, phospholipids and Quillaja saponins under a specific stoichiometry. The complex displays immune stimulating properties and is thus mainly used as a vaccine adjuvant in order to induce a stronger immune response and longer protection. A specific adjuvant based on ISCOM technology is Matrix-M.
An antiviral stockpile is a reserve supply of essential antiviral medications in case of shortage. Many countries have chosen to stockpile antiviral medications against pandemic influenza. Because of the time required to prepare and distribute an influenza vaccine, these stockpiles are the only medical defense against widespread infection for the first six months. The stockpiles may be in the form of capsules or simply as the active pharmaceutical ingredient, which is stored in sealed drums and, when needed, dissolved in water to make a bitter-tasting, clear liquid.

Albertus Dominicus Marcellinus Erasmus "Ab" Osterhaus is a leading Dutch virologist and influenza expert. An Emeritus Professor of Virology at Erasmus University Rotterdam since 1993, Osterhaus is known throughout the world for his work on SARS and H5N1, the pathogen that causes avian influenza.

Betacoronavirus cameli, or EMC/2012 (HCoV-EMC/2012), is the virus that causes Middle East respiratory syndrome (MERS). It is a species of coronavirus which infects humans, bats, and camels. The infecting virus is an enveloped, positive-sense, single-stranded RNA virus which enters its host cell by binding to the DPP4 receptor. The species is a member of the genus Betacoronavirus and subgenus Merbecovirus.
Novel coronavirus (nCoV) is a provisional name given to coronaviruses of medical significance before a permanent name is decided upon. Although coronaviruses are endemic in humans and infections normally mild, such as the common cold, cross-species transmission has produced some unusually virulent strains which can cause viral pneumonia and in serious cases even acute respiratory distress syndrome and death.
Pipistrellus bat coronavirus HKU5 is an enveloped, positive-sense single-stranded RNA mammalian Group 2 Betacoronavirus discovered in Japanese Pipistrellus in Hong Kong. This strain of coronavirus is closely related to the newly identified novel MERS-CoV that is responsible for the 2012 Middle East respiratory syndrome-related coronavirus outbreaks in Saudi Arabia, Jordan, United Arab Emirates, the United Kingdom, France, and Italy.

Betacoronavirus hongkonense is a species of coronavirus in humans and animals. It causes an upper respiratory disease with symptoms of the common cold, but can advance to pneumonia and bronchiolitis. It was first discovered in January 2004 from one man in Hong Kong. Subsequent research revealed it has global distribution and earlier genesis.

Alphacoronavirus chicagoense is a species of coronavirus which infects humans and bats. It is an enveloped, positive-sense, single-stranded RNA virus which enters its host cell by binding to the APN receptor. Along with Human coronavirus OC43, it is one of the viruses responsible for the common cold. HCoV-229E is a member of the genus Alphacoronavirus and subgenus Duvinacovirus.
Tylonycteris bat coronavirus HKU4 is an enveloped, positive-sense single-stranded RNA virus mammalian Group 2 Betacoronavirus that has been found to be genetically related to the Middle East respiratory syndrome-related coronavirus (MERS-CoV) that is responsible for the 2012 Middle East respiratory syndrome coronavirus outbreak in Saudi Arabia, Jordan, United Arab Emirates, the United Kingdom, France, and Italy.
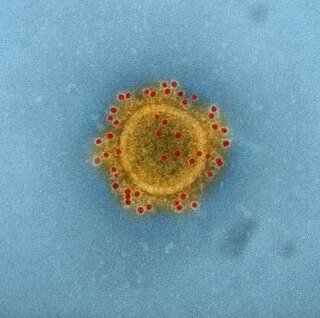
MERS coronavirus EMC/2012 is a strain of coronavirus isolated from the sputum of the first person to become infected with what was later named Middle East respiratory syndrome–related coronavirus (MERS-CoV), a virus that causes Middle East respiratory syndrome (MERS).
Biotechnology risk is a form of existential risk from biological sources, such as genetically engineered biological agents. The release of such high-consequence pathogens could be

Maria Petronella Gerarda Koopmans is a Dutch virologist who is Head of the Erasmus MC Department of Viroscience. Her research considers emerging infectious diseases, noroviruses and veterinary medicine. In 2018 she was awarded the Netherlands Organisation for Scientific Research (NWO) Stevin Prize. She serves on the scientific advisory group of the World Health Organization.
Gain-of-function research is medical research that genetically alters an organism in a way that may enhance the biological functions of gene products. This may include an altered pathogenesis, transmissibility, or host range, i.e., the types of hosts that a microorganism can infect. This research is intended to reveal targets to better predict emerging infectious diseases and to develop vaccines and therapeutics. For example, influenza B can infect only humans and harbor seals. Introducing a mutation that would allow influenza B to infect rabbits in a controlled laboratory situation would be considered a gain-of-function experiment, as the virus did not previously have that function. That type of experiment could then help reveal which parts of the virus's genome correspond to the species that it can infect, enabling the creation of antiviral medicines which block this function.













