
Rotavirus is a genus of double-stranded RNA viruses in the family Reoviridae. Rotaviruses are the most common cause of diarrhoeal disease among infants and young children. Nearly every child in the world is infected with a rotavirus at least once by the age of five. Immunity develops with each infection, so subsequent infections are less severe. Adults are rarely affected. There are nine species of the genus, referred to as A, B, C, D, F, G, H, I and J. Rotavirus A, the most common species, causes more than 90% of rotavirus infections in humans.

In molecular genetics, the three prime untranslated region (3′-UTR) is the section of messenger RNA (mRNA) that immediately follows the translation termination codon. The 3′-UTR often contains regulatory regions that post-transcriptionally influence gene expression.

An enterotoxin is a protein exotoxin released by a microorganism that targets the intestines.
The 5′ untranslated region is the region of a messenger RNA (mRNA) that is directly upstream from the initiation codon. This region is important for the regulation of translation of a transcript by differing mechanisms in viruses, prokaryotes and eukaryotes. While called untranslated, the 5′ UTR or a portion of it is sometimes translated into a protein product. This product can then regulate the translation of the main coding sequence of the mRNA. In many organisms, however, the 5′ UTR is completely untranslated, instead forming complex secondary structure to regulate translation.
An internal ribosome entry site, abbreviated IRES, is an RNA element that allows for translation initiation in a cap-independent manner, as part of the greater process of protein synthesis. In eukaryotic translation, initiation typically occurs at the 5' end of mRNA molecules, since 5' cap recognition is required for the assembly of the initiation complex. The location for IRES elements is often in the 5'UTR, but can also occur elsewhere in mRNAs.
Eukaryotic translation is the biological process by which messenger RNA is translated into proteins in eukaryotes. It consists of four phases: gene translation, elongation, termination, and recapping.

Alfalfa mosaic virus (AMV), also known as Lucerne mosaic virus or Potato calico virus, is a worldwide distributed phytopathogen that can lead to necrosis and yellow mosaics on a large variety of plant species, including commercially important crops. It is the only Alfamovirus of the family Bromoviridae. In 1931 Weimer J.L. was the first to report AMV in alfalfa. Transmission of the virus occurs mainly by some aphids, by seeds or by pollen to the seed.
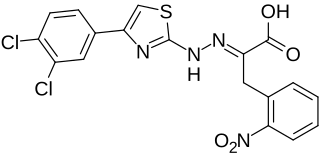
4EGI-1 is a synthetic chemical compound which has been found to interfere with the growth of certain types of cancer cells in vitro. Its mechanism of action involves interruption of the binding of cellular initiation factor proteins involved in the translation of transcribed mRNA at the ribosome. The inhibition of these initiation factors prevents the initiation and translation of many proteins whose functions are essential to the rapid growth and proliferation of cancer cells.
Poly(A)-binding protein is a RNA-binding protein which triggers the binding of eukaryotic initiation factor 4 complex (eIF4G) directly to the poly(A) tail of mRNA which is 200-250 nucleotides long. The poly(A) tail is located on the 3' end of mRNA and was discovered by Mary Edmonds, who also characterized the poly-A polymerase enzyme that generates the poly(a) tail. The binding protein is also involved in mRNA precursors by helping polyadenylate polymerase add the poly(A) nucleotide tail to the pre-mRNA before translation. The nuclear isoform selectively binds to around 50 nucleotides and stimulates the activity of polyadenylate polymerase by increasing its affinity towards RNA. Poly(A)-binding protein is also present during stages of mRNA metabolism including nonsense-mediated decay and nucleocytoplasmic trafficking. The poly(A)-binding protein may also protect the tail from degradation and regulate mRNA production. Without these two proteins in-tandem, then the poly(A) tail would not be added and the RNA would degrade quickly.
A ribosome binding site, or ribosomal binding site (RBS), is a sequence of nucleotides upstream of the start codon of an mRNA transcript that is responsible for the recruitment of a ribosome during the initiation of translation. Mostly, RBS refers to bacterial sequences, although internal ribosome entry sites (IRES) have been described in mRNAs of eukaryotic cells or viruses that infect eukaryotes. Ribosome recruitment in eukaryotes is generally mediated by the 5' cap present on eukaryotic mRNAs.

RoXaN also known as ZC3H7B, is a protein that in humans is encoded by the ZC3H7B gene. RoXaN is a protein that contains tetratricopeptide repeat and leucine-aspartate repeat as well as zinc finger domains. This protein also interacts with the rotavirus non-structural protein NSP3.
NSP1 (NS53), the product of rotavirus gene 5, is a nonstructural RNA-binding protein that contains a cysteine-rich region and is a component of early replication intermediates. RNA-folding predictions suggest that this region of the NSP1 mRNA can interact with itself, producing a stem-loop structure similar to that found near the 5'-terminus of the NSP1 mRNA.

NSP2 (NS35), is a rotavirus nonstructural RNA-binding protein that accumulates in cytoplasmic inclusions (viroplasms) and is required for genome replication. NSP2 is closely associated in vivo with the viral replicase. The non-structural protein NSP5 plays a role in the structure of viroplasms mediated by its interaction with NSP2.

Rotavirus protein NSP3 (NS34) is bound to the 3' end consensus sequence of viral mRNAs in infected cells.
NSP5 encoded by genome segment 11 of group A rotaviruses. In virus-infected cells NSP5 accumulates in the viroplasms. NSP5 has been shown to be autophosphorylated. Interaction of NSP5 with NSP2 was also demonstrated. In rotavirus-infected cells, the non-structural proteins NSP5 and NSP2 localize in complexes called viroplasms, where replication and assembly occur and they can drive the formation of viroplasm-like structures in the absence of other rotaviral proteins and rotavirus replication.

Polyadenylate-binding protein 1 is a protein that in humans is encoded by the PABPC1 gene. The protein PABP1 binds mRNA and facilitates a variety of functions such as transport into and out of the nucleus, degradation, translation, and stability. There are two separate PABP1 proteins, one which is located in the nucleus (PABPN1) and the other which is found in the cytoplasm (PABPC1). The location of PABP1 affects the role of that protein and its function with RNA.

Eukaryotic translation initiation factor 4 gamma 1 is a protein that in humans is encoded by the EIF4G1 gene.

Eukaryotic initiation factor 4A-I is a 46 kDa cytosolic protein that, in humans, is encoded by the EIF4A1 gene, which is located on chromosome 17. It is the most prevalent member of the eIF4A family of ATP-dependant RNA helicases, and plays a critical role in the initiation of cap-dependent eukaryotic protein translation as a component of the eIF4F translation initiation complex. eIF4A1 unwinds the secondary structure of RNA within the 5'-UTR of mRNA, a critical step necessary for the recruitment of the 43S preinitiation complex, and thus the translation of protein in eukaryotes. It was first characterized in 1982 by Grifo, et al., who purified it from rabbit reticulocyte lysate.
Eukaryotic translation initiation factor 4 G (eIF4G) is a protein involved in eukaryotic translation initiation and is a component of the eIF4F cap-binding complex. Orthologs of eIF4G have been studied in multiple species, including humans, yeast, and wheat. However, eIF4G is exclusively found in domain Eukarya, and not in domains Bacteria or Archaea, which do not have capped mRNA. As such, eIF4G structure and function may vary between species, although the human EIF4G1 has been the focus of extensive studies.

Eukaryotic initiation factor 4F (eIF4F) is a heterotrimeric protein complex that binds the 5' cap of messenger RNAs (mRNAs) to promote eukaryotic translation initiation. The eIF4F complex is composed of three non-identical subunits: the DEAD-box RNA helicase eIF4A, the cap-binding protein eIF4E, and the large "scaffold" protein eIF4G. The mammalian eIF4F complex was first described in 1983, and has been a major area of study into the molecular mechanisms of cap-dependent translation initiation ever since.













