
In molecular biology, a transcription factor (TF) is a protein that controls the rate of transcription of genetic information from DNA to messenger RNA, by binding to a specific DNA sequence. The function of TFs is to regulate—turn on and off—genes in order to make sure that they are expressed in the desired cells at the right time and in the right amount throughout the life of the cell and the organism. Groups of TFs function in a coordinated fashion to direct cell division, cell growth, and cell death throughout life; cell migration and organization during embryonic development; and intermittently in response to signals from outside the cell, such as a hormone. There are up to 1600 TFs in the human genome. Transcription factors are members of the proteome as well as regulome.
In biology, a sequence motif is a nucleotide or amino-acid sequence pattern that is widespread and usually assumed to be related to biological function of the macromolecule. For example, an N-glycosylation site motif can be defined as Asn, followed by anything but Pro, followed by either Ser or Thr, followed by anything but Pro residue.
A document-term matrix is a mathematical matrix that describes the frequency of terms that occur in a collection of documents. In a document-term matrix, rows correspond to documents in the collection and columns correspond to terms. This matrix is a specific instance of a document-feature matrix where "features" may refer to other properties of a document besides terms. It is also common to encounter the transpose, or term-document matrix where documents are the columns and terms are the rows. They are useful in the field of natural language processing and computational text analysis.
RNA polymerase 1 is, in higher eukaryotes, the polymerase that only transcribes ribosomal RNA, a type of RNA that accounts for over 50% of the total RNA synthesized in a cell.
A position weight matrix (PWM), also known as a position-specific weight matrix (PSWM) or position-specific scoring matrix (PSSM), is a commonly used representation of motifs (patterns) in biological sequences.
The MADS box is a conserved sequence motif. The genes which contain this motif are called the MADS-box gene family. The MADS box encodes the DNA-binding MADS domain. The MADS domain binds to DNA sequences of high similarity to the motif CC[A/T]6GG termed the CArG-box. MADS-domain proteins are generally transcription factors. The length of the MADS-box reported by various researchers varies somewhat, but typical lengths are in the range of 168 to 180 base pairs, i.e. the encoded MADS domain has a length of 56 to 60 amino acids. There is evidence that the MADS domain evolved from a sequence stretch of a type II topoisomerase in a common ancestor of all extant eukaryotes.
The chicken ovalbumin upstream promoter transcription factor (COUP-TFs) proteins are members of the nuclear receptor family of intracellular transcription factors. There are two variants of the COUP-TFs, labeled as COUP-TFI and COUP-TFII encoded by the NR2F1 and NR2F2 genes respectively.
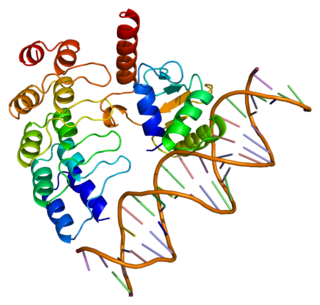
GA-binding protein alpha chain is a protein that in humans is encoded by the GABPA gene.

COUP-TF1 also known as NR2F1 is a protein that in humans is encoded by the NR2F1 gene. This protein is a member of nuclear hormone receptor family of steroid hormone receptors.

COUP-TFII, also known as NR2F2 is a protein that in humans is encoded by the NR2F2 gene. The COUP acronym stands for chicken ovalbumin upstream promoter.

Pho4 is a protein with a basic helix-loop-helix (bHLH) transcription factor. It is found in S. cerevisiae and other yeasts. It functions as a transcription factor to regulate phosphate responsive genes located in yeast cells. The Pho4 protein homodimer is able to do this by binding to DNA sequences containing the bHLH binding site 5'-CACGTG-3'. This sequence is found in the promoters of genes up-regulated in response to phosphate availability such as the PHO5 gene.
DNA binding sites are a type of binding site found in DNA where other molecules may bind. DNA binding sites are distinct from other binding sites in that (1) they are part of a DNA sequence and (2) they are bound by DNA-binding proteins. DNA binding sites are often associated with specialized proteins known as transcription factors, and are thus linked to transcriptional regulation. The sum of DNA binding sites of a specific transcription factor is referred to as its cistrome. DNA binding sites also encompasses the targets of other proteins, like restriction enzymes, site-specific recombinases and methyltransferases.
RegulonDB is a database of the regulatory network of gene expression in Escherichia coli K-12. RegulonDB also models the organization of the genes in transcription units, operons and regulons. A total of 120 sRNAs with 231 total interactions which all together regulate 192 genes are also included. RegulonDB was founded in 1998 and also contributes data to the EcoCyc database.
YEASTRACT is a curated repository of more than 48000 regulatory associations between transcription factors (TF) and target genes in Saccharomyces cerevisiae, based on more than 1200 bibliographic references. It also includes the description of about 300 specific DNA binding sites for more than a hundred characterized TFs. Further information about each Yeast gene has been extracted from the Saccharomyces Genome Database (SGD). For each gene the associated Gene Ontology (GO) terms and their hierarchy in GO was obtained from the GO consortium. Currently, YEASTRACT maintains more than 7100 terms from GO. The nucleotide sequences of the promoter and coding regions for Yeast genes were obtained from Regulatory Sequence Analysis Tools (RSAT). All the information in YEASTRACT is updated regularly to match the latest data from SGD, GO consortium, RSA Tools and recent literature on yeast regulatory networks.
TRANSFAC is a manually curated database of eukaryotic transcription factors, their genomic binding sites and DNA binding profiles. The contents of the database can be used to predict potential transcription factor binding sites.
YeTFaSCo is a database of transcription factors for Saccharomyces cerevisiae.

Myelin regulatory factor, also known as myelin gene regulatory factor (MRF), is a protein that in humans is encoded by the MYRF gene.

Gary Stormo is an American geneticist and currently Joseph Erlanger Professor in the Department of Genetics and the Center for Genome Sciences and Systems Biology at Washington University School of Medicine in St Louis. He is considered one of the pioneers of bioinformatics and genomics. His research combines experimental and computational approaches in order to identify and predict regulatory sequences in DNA and RNA, and their contributions to the regulatory networks that control gene expression.
CollecTF is a database of transcription factor binding sites in the Bacteria domain.
JASPAR is an open access and widely used database of manually curated, non-redundant transcription factor (TF) binding profiles stored as position frequency matrices (PFM) and transcription factor flexible models (TFFM) for TFs from species in six taxonomic groups. From the supplied PFMs, users may generate position-specific weight matrices (PWM). The JASPAR database was introduced in 2004. There were seven major updates and new releases in 2006, 2008, 2010, 2014, 2016, 2018, 2020 and 2022, which is the latest release of JASPAR.






